













| General Information: |
|
| Name(s) found: |
RGA2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1009 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSADPINDQS SLCVRCNKSI ASSQVYELES KKWHDQCFTC YKCDKKLNAD SDFLVLDIGT 60
61 LICYDCSDKC TNCGDKIDDT AIILPSSNEA YCSNCFRCCR CSNRIKNLKY AKTKRGLCCM 120
121 DCHEKLLRKK QLLLENQTKN SSKEDFPIKL PERSVKRPLS PTRINGKSDV STNNTAISKN 180
181 LVSSNEDQQL TPQVLVSQER DESSLNDNND NDNSKDREET SSHARTVSID DILNSTLEHD 240
241 SNSIEEQSLV DNEDYINKMG EDVTYRLLKP QRANRDSIVV KDPRIPNSNS NANRFFSIYD 300
301 KEETDKDDTD NKENEIIVNT PRNSTDKITS PLNSPMAVQM NEEVEPPHGL ALTLSEATKE 360
361 NNKSSQGIQT STSKSMNHVS PITRTDTVEM KTSTSSSTLR LSDNGSFSRP QTADNLLPHK 420
421 KVAPSPNKKL SRSFSLKSKN FVHNLKSKTS EMLDPKHPHH STSIQESDTH SGWGVSSTHT 480
481 NIRKSKAKKN PVSRGQSDST IYNTLPQHGN FTVPEFNHKK AQSSLGSISK KQNSNDTATN 540
541 RRINGSFTSS SSGHHIAMFR TPPLESGPLF KRPSLSSESA HHRSSSLQTS RSTNALLEDD 600
601 STKVDATDES ATSLEKDFYF TELTLRKLKL DVRELEGTKK KLLQDVENLR LAKERLLNDV 660
661 DNLTREKDKQ SASSRESLEQ KENIATSITV KSPSSNSDRK GSISNASPKP RFWKIFSSAK 720
721 DHQVGDLESQ QRSPNSSSGG TTNIAQKEIS SPKLIRVHDE LPSPGKVPLS PSPKRLDYTP 780
781 DGSHLYGSSL QARCAYEKST VPIIIRCCID RIEKDDIGLN MEGLYRKSGS QTLVEEIENE 840
841 FAQNNSLHSD TLSPKLNALL NQDIHAVASV LKRYLRKLPD PVLSFSIYDA LIDLVRNNQL 900
901 IERLPLNNDK FLDSPQKVTI YEMVLKSLLE IFKILPVEHQ EVLKVLAAHI GKVRRCSERN 960
961 LMNLHNLSLV FAPSLIHDFD GEKDIVDMKE RNYIVEFILG NYRDIFKQA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
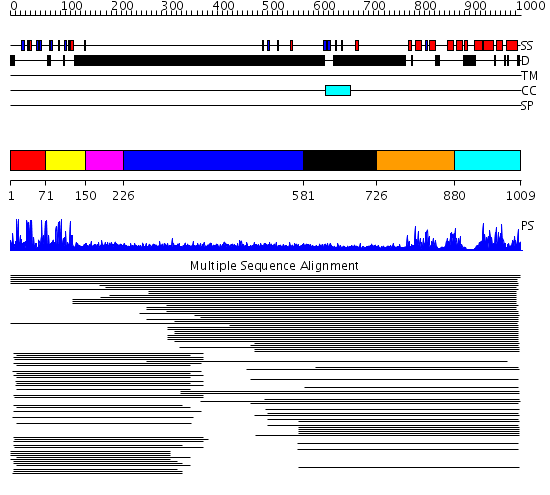
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | 38.228787 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 2 | View Details | [71..149] | 38.228787 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 3 | View Details | [150..225] | 10.154902 | Insulin gene enhancer protein isl-1 | |
| 4 | View Details | [226..580] | 3.138996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [581..725] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [726..879] | 193.30103 | p50 RhoGAP domain | |
| 7 | View Details | [880..1009] | 193.30103 | p50 RhoGAP domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)