













| General Information: |
|
| Name(s) found: |
PRK1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 810 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTPQISLYE PGTILTVGSH HAKIIKYLTS GGFAQVYTAE ISPPDPYSNA NIACLKRVIV 60
61 PHKQGLNTLR AEVDAMKLLR NNKHVVSYID SHAARSVNGI AYEVFVLMEF CERGGLIDFM 120
121 NTRLQNRLQE SEILEIMSQT VQGITAMHAL QPPLIHRDIK IENVLISHDG LYKVCDFGSV 180
181 SGVIRPPRNT QEFNYVQHDI LTNTTAQYRS PEMIDLYRGL PIDEKSDIWA LGVFLYKICY 240
241 YTTPFEKSGE AGILHARYQY PSFPQYSDRL KNLIRLMLME APSQRPNICQ VLEEVSRLQN 300
301 KPCPIRNFYL LRAMNQNANT QLAGEPSSTT YVPTQKFIPV QSLQSINQPP NMMPVTHVST 360
361 TPNLGTFPIS INDNNKTEVT AHAGLQVGSH SNLTSPLMKT KSVPLSDEFA SLYYKELHPF 420
421 QKSQTFKSVE SFQSPQRKSM PPLSLTPVNN DIFDRVSAIN RPNNYVDSET QTIDNMAVPN 480
481 LKLSPTITSK SLSSTKEIAA PDNINGSKIV RSLSSKLKKV ITGESRGNSP IKSRQNTGDS 540
541 IRSAFGKLRH GFTGNSVNNS RSASFDNNNV NGNGNNTNRR LVSSSTSSFP KFNSDTKRKE 600
601 ESDKNQRLEK RRSMPPSILS DFDQHERNNS RTGSRDYYRS HSPVKKTQAS AKTTSKPTLI 660
661 PDNGNVNINQ EKKESIQRRV HNLLKSSDDP VTYKSASGYG KYTDIGTETS NRHSSVRITP 720
721 ITEEKFKKTL KDGVLDIKTK SNGKDKSRPP RPPPKPLHLR TEIQKIRNFS RLQSKKLPIE 780
781 RISSEATETI VDVNVDDLEA DFRKRFPSKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
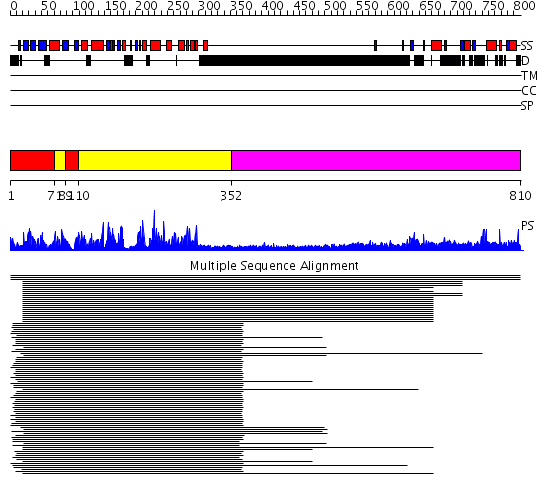
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] [89..109] |
222.201249 | gamma-subunit of glycogen phosphorylase kinase (Phk) | |
| 2 | View Details | [71..88] [110..351] |
222.201249 | gamma-subunit of glycogen phosphorylase kinase (Phk) | |
| 3 | View Details | [352..810] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [709..810] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)