













| General Information: |
|
| Name(s) found: |
PPZ1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 692 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNSSSKSSK KDSHSNSSSR NPRPQVSRTE TSHSVKSAKS NKSSRSRRSL PSSSTTNTNS 60
61 NVPDPSTPSK PNLEVNHQRH SSHTNRYHFP SSSHSHSNSQ NELLTTPSSS STKRPSTSRR 120
121 SSYNTKAAAD LPPSMIQMEP KSPILKTNNS STHVSKHKSS YSSTYYENAL TDDDNDDKDN 180
181 DISHTKRFSR SSNSRPSSIR SGSVSRRKSD VTHEEPNNGS YSSNNQENYL VQALTRSNSH 240
241 ASSLHSRKSS FGSDGNTAYS TPLNSPGLSK LTDHSGEYFT SNSTSSLNHH SSRDIYPSKH 300
301 ISNDDDIENS SQLSNIHASM ENVNDKNNNI TDSKKDPNEE FNDIMQSSGN KNAPKKFKKP 360
361 IDIDETIQKL LDAGYAAKRT KNVCLKNNEI LQICIKAREI FLSQPSLLEL SPPVKIVGDV 420
421 HGQYGDLLRL FTKCGFPPSS NYLFLGDYVD RGKQSLETIL LLFCYKIKYP ENFFLLRGNH 480
481 ECANVTRVYG FYDECKRRCN IKIWKTFIDT FNTLPLAAIV AGKIFCVHGG LSPVLNSMDE 540
541 IRHVVRPTDV PDFGLINDLL WSDPTDSPNE WEDNERGVSY CYNKVAINKF LNKFGFDLVC 600
601 RAHMVVEDGY EFFNDRSLVT VFSAPNYCGE FDNWGAVMSV SEGLLCSFEL LDPLDSAALK 660
661 QVMKKGRQER KLANQQQQMM ETSITNDNES QQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
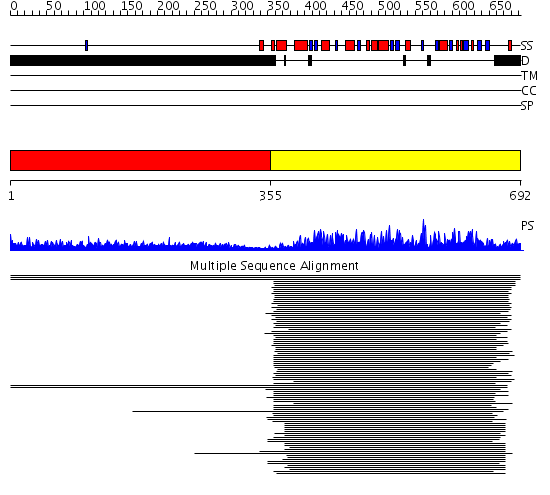
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | 13.84 | Sec24 | |
| 2 | View Details | [355..692] | 1409.0 | Protein phosphatase-1 (PP-1) | |
| 3 | View Details | [354..692] | 161.0 | No description for 2o8aA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)