













| General Information: |
|
| Name(s) found: |
PCF11_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 626 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDHDTEVIVK DFNSILEELT FNSRPIITTL TKLAEENISC AQYFVDAIES RIEKCMPKQK 60
61 LYAFYALDSI CKNVGSPYTI YFSRNLFNLY KRTYLLVDNT TRTKLINMFK LWLNPNDTGL 120
121 PLFEGSALEK IEQFLIKASA LHQKNLQAML PTPTVPLLLR DIDKLTCLTS ERLKNQPNDE 180
181 KLKMKLLVLS QLKQELKREK LTLNALKQVQ MQLRQVFSQD QQVLQERMRY HELQQQQQQQ 240
241 QQQQQQQQQQ QQQYHETKDM VGSYTQNSNS AIPLFGNNSD TTNQQNSLSS SLFGNISGVE 300
301 SFQEIEKKKS LNKINNLYAS LKAEGLIYTP PKESIVTLYK KLNGHSNYSL DSHEKQLMKN 360
361 LPKIPLLNDI LSDCKAYFAT VNIDVLNNPS LQLSEQTLLQ ENPIVQNNLI HLLYRSKPNK 420
421 CSVCGKRFGN SESEKLLQNE HLDWHFRINT RIKGSQNTAN TGISNSNLNT TTTRKNIQSR 480
481 NWYLSDSQWA AFKDDEITST KHKNDYTDPH ANKNIDKSAL NIHADENDEG SVDNTLGSDR 540
541 SNELEIRGKY VVVPETSQDM AFKCPICKET VTGVYDEESG EWVWKNTIEV NGKYFHSTCY 600
601 HETSQNSSKS NSGKVGLDDL KKLVTK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
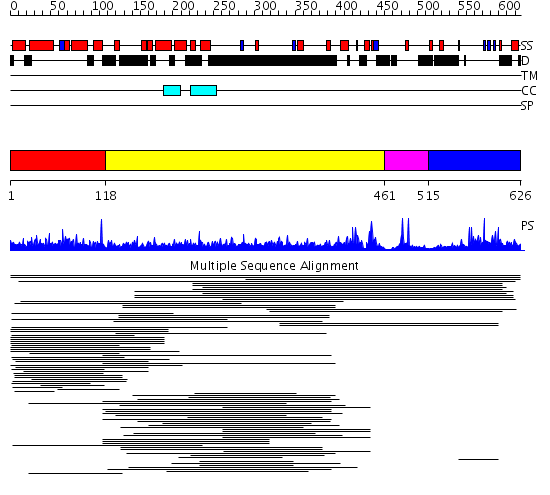
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 5.03998 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [118..460] | 19.057997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [461..514] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [515..626] | 1.013996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)