













| General Information: |
|
| Name(s) found: |
PAP2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 584 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGAKSVTASS SKKIKNRHNG KVKKSKKIKK VRKPQKSISL NDENEVEILP SRNEQETNKL 60
61 PKDHVTADGI LVLEHKSDDD EGFDVYDGHF DNPTDIPSTT EESKTPSLAV HGDEKDLANN 120
121 DDFISLSASS EDEQAEQEEE REKQELEIKK EKQKEILNTD YPWILNHDHS KQKEISDWLT 180
181 FEIKDFVAYI SPSREEIEIR NQTISTIREA VKQLWPDADL HVFGSYSTDL YLPGSDIDCV 240
241 VTSELGGKES RNNLYSLASH LKKKNLATEV EVVAKARVPI IKFVEPHSGI HIDVSFERTN 300
301 GIEAAKLIRE WLDDTPGLRE LVLIVKQFLH ARRLNNVHTG GLGGFSIICL VFSFLHMHPR 360
361 IITNEIDPKD NLGVLLIEFF ELYGKNFGYD DVALGSSDGY PVYFPKSTWS AIQPIKNPFS 420
421 LAIQDPGDES NNISRGSFNI RDIKKAFAGA FDLLTNRCFE LHSATFKDRL GKSILGNVIK 480
481 YRGKARDFKD ERGLVLNKAI IENENYHKKR SRIIHDEDFA EDTVTSTATA TTTDDDYEIT 540
541 NPPAKKAKIE EKPESEPAKR NSGETYITVS SEDDDEDGYN PYTL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
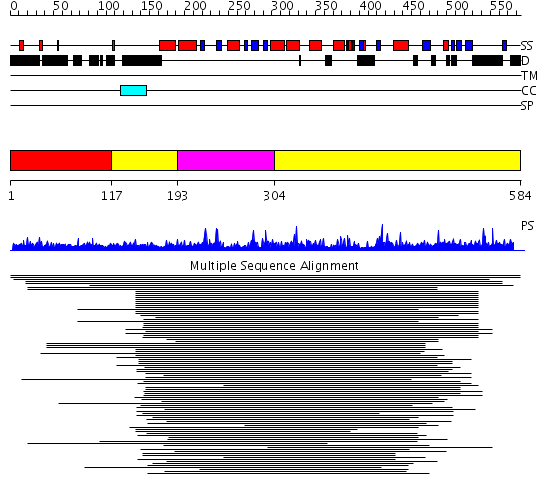
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..192] [304..584] |
91.0 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain | |
| 3 | View Details | [193..303] | 91.0 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.25 |
Source: Reynolds et al. (2008)