













| General Information: |
|
| Name(s) found: |
NGR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 672 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMSNVANASQ RQENPYIIPL PPSSTVETST EPPRTLWMGD LDPSFDEATI EEIWSKLDKK 60
61 VIVKLIRAKK NLLIPCSSTS SSNNNTSEEN AENQQSASNS TDQLDNSQMI NINGISFIDP 120
121 STTQLHHAGY CFVEFETQKD AKFALSLNAT PLPNFYSPTT NSQTNPTFKR TFRLNWASGA 180
181 TLQSSIPSTP EFSLFVGDLS PTATEADLLS LFQTRFKSVK TVRVMTDPLT GSSRCFGFVR 240
241 FGDEDERRRA LIEMSGKWFQ GRALRVAYAT PRNNMMLQLQ EQQQQQQQLQ QQHQQLDQED 300
301 NNGPLLIKTA NNLIQNNSNM LPLNALHNAP PMHLNEGGIS NMRVNDSLPS NTYNTDPTNT 360
361 TVFVGGLVPK TTEFQLRSLF KPFGPILNVR IPNGKNCGFV KFEKRIDAEA SIQGLQGFIV 420
421 GGSPIRLSWG RPSSSNAKTN STIMGASQYM SSNGLRAPSA ASSVDNSKQI LEQYAEDKRR 480
481 LFLHQQQQQQ QQQQQDGNFS MEQMAHNNYY NYNNYDYHRN KNGSHSDLVN LQRSNVPYMQ 540
541 EDGALYPHQY SSPSYSLHPT GNQFSNATNN LPQFGNAMSI SMQLPNGNSN KTASSMNTNP 600
601 NTNMIMNSNM NMNMNVNPVP YGMGNGANMY DVSRMMTPPL NIAPNSNNSK SSIMNKHPNR 660
661 NNVPPIHPSL LH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
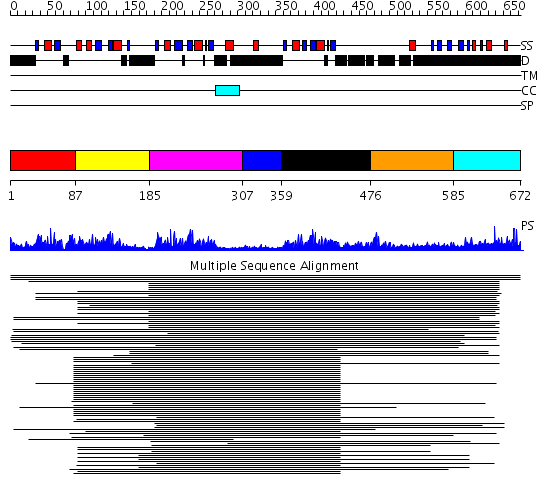
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 92.54902 | Poly(A)-binding protein | |
| 2 | View Details | [87..184] | 89.28043 | Hu antigen D (Hud) | |
| 3 | View Details | [185..306] | 89.28043 | Hu antigen D (Hud) | |
| 4 | View Details | [307..358] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [359..475] | 13.221849 | CBP20, 20KDa nuclear cap-binding protein | |
| 6 | View Details | [476..584] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [585..672] | 11.0 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)