













| General Information: |
|
| Name(s) found: |
NGL3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 505 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSQVEGKIS PSQKESSSTS GLVSPSEDGP AHQKIHRDQL SVDQIKKIRE ERAQKRQVRR 60
61 NSLISQGKDP DFPTPDLQFI ERPFLPINHD NSKGLTPATI QVTQDSLDVK IMTYNTLAQT 120
121 LIRRDFFPES GPALKWHKRS KVLVHELKKY RPDVVSLQEV DYNELNFWQE NFHKLGFDVI 180
181 FKRHEGKTHG LLVAWNNKKF QLDNDWMLDY DNILAGNVIS ARTRTKNIAL IISLYFKGIT 240
241 DSSSRGIIVA NTHLFWHPFG VFERLRQSYL VLQKIQEIKA CSKYNGWHSL LMGDFNTEPE 300
301 EPPYLAITKR PLILKGPIRA MVECSLAYRY SKKRNGEESD QDDEECDEKS RGEGHSDQPQ 360
361 NPKPESFTAT KEEKALVNQL VALHNSLHVK GVSLYGIGYG KVHPENANGS HGEPGLSNWA 420
421 NTWCGLLDYI FYIEGDHNQD TRQKEPLNAF EGNNNVKIIG YLRMPCAQEM PKHSQPFEGE 480
481 YASDHISLMC QIRLFFGGEK VHSLK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
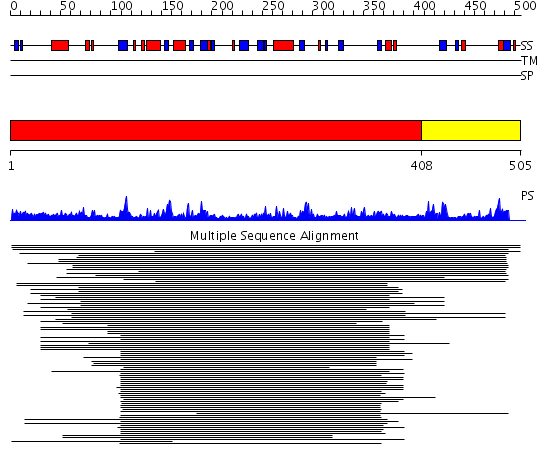
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..407] | 50.0 | DNA repair endonuclease Hap1 | |
| 2 | View Details | [408..505] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)