













| General Information: |
|
| Name(s) found: |
METE_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQSAVLGFP RIGPNRELKK ATEGYWNGKI TVDELFKVGK DLRTQNWKLQ KEAGVDIIPS 60
61 NDFSFYDQVL DLSLLFNVIP DRYTKYDLSP IDTLFAMGRG LQRKATETEK AVDVTALEMV 120
121 KWFDSNYHYV RPTFSKTTQF KLNGQKPVDE FLEAKELGIH TRPVLLGPVS YLFLGKADKD 180
181 SLDLEPLSLL EQLLPLYTEI LSKLASAGAT EVQIDEPVLV LDLPANAQAA IKKAYTYFGE 240
241 QSNLPKITLA TYFGTVVPNL DAIKGLPVAA LHVDFVRAPE QFDEVVAAIG NKQTLSVGIV 300
301 DGRNIWKNDF KKSSAIVNKA IEKLGADRVV VATSSSLLHT PVDLNNETKL DAEIKGFFSF 360
361 ATQKLDEVVV ITKNVSGQDV AAALEANAKS VESRGKSKFI HDAAVKARVA SIDEKMSTRA 420
421 APFEQRLPEQ QKVFNLPLFP TTTIGSFPQT KDIRINRNKF NKGTISAEEY EKFINSEIEK 480
481 VIRFQEEIGL DVLVHGEPER NDMVQYFGEQ INGYAFTVNG WVQSYGSRYV RPPIIVGDLS 540
541 RPKAMSVKES VYAQSITSKP VKGMLTGPIT CLRWSFPRDD VDQKTQAMQL ALALRDEVND 600
601 LEAAGIKVIQ VDEPALREGL PLREGTERSA YYTWAAEAFR VATSGVANKT QIHSHFCYSD 660
661 LDPNHIKALD ADVVSIEFSK KDDANYIAEF KNYPNHIGLG LFDIHSPRIP SKDEFIAKIS 720
721 TILKSYPAEK FWVNPDCGLK TRGWEETRLS LTHMVEAAKY FREQYKN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
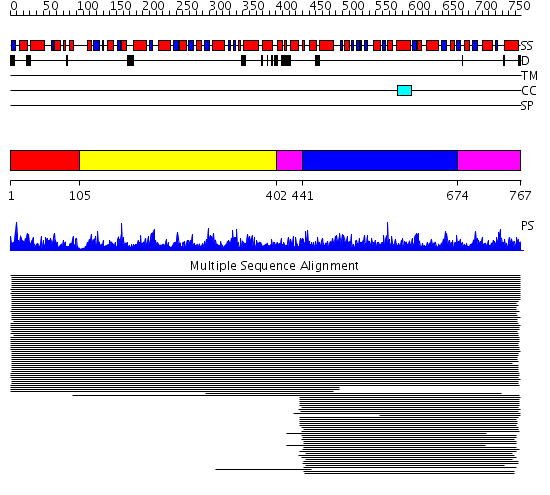
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..104] | 1.042975 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [105..401] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [402..440] [674..767] |
11.96 | Uroporphyrinogen decarboxylase, UROD | |
| 4 | View Details | [441..673] | 11.96 | Uroporphyrinogen decarboxylase, UROD |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)