













| General Information: |
|
| Name(s) found: |
KICH_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 582 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQESRPGSV RSYSVGYQAR SRSSSQRRHS LTRQRSSQRL IRTISIESDV SNITDDDDLR 60
61 AVNEGVAGVQ LDVSETANKG PRRASATDVT DSLGSTSSEY IEIPFVKETL DASLPSDYLK 120
121 QDILNLIQSL KISKWYNNKK IQPVAQDMNL VKISGAMTNA IFKVEYPKLP SLLLRIYGPN 180
181 IDNIIDREYE LQILARLSLK NIGPSLYGCF VNGRFEQFLE NSKTLTKDDI RNWKNSQRIA 240
241 RRMKELHVGV PLLSSERKNG SACWQKINQW LRTIEKVDQW VGDPKNIENS LLCENWSKFM 300
301 DIVDRYHKWL ISQEQGIEQV NKNLIFCHND AQYGNLLFTA PVMNTPSLYT APSSTSLTSQ 360
361 SSSLFPSSSN VIVDDIINPP KQEQSQDSKL VVIDFEYAGA NPAAYDLANH LSEWMYDYNN 420
421 AKAPHQCHAD RYPDKEQVLN FLYSYVSHLR GGAKEPIDEE VQRLYKSIIQ WRPTVQLFWS 480
481 LWAILQSGKL EKKEASTAIT REEIGPNGKK YIIKTEPESP EEDFVENDDE PEAGVSIDTF 540
541 DYMAYGRDKI AVFWGDLIGL GIITEEECKN FSSFKFLDTS YL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
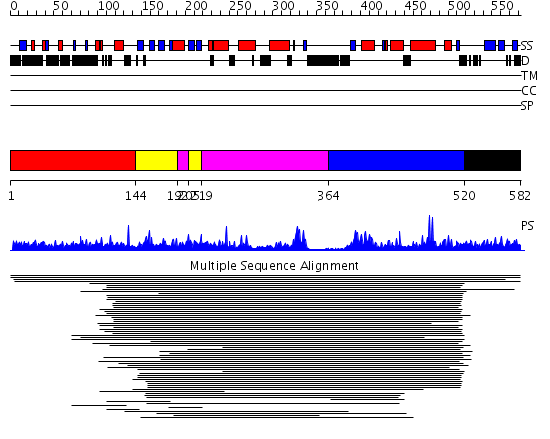
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [144..191] [205..218] |
12.95 | Type IIIa 3',5"-aminoglycoside phosphotransferase | |
| 3 | View Details | [192..204] [219..363] |
12.95 | Type IIIa 3',5"-aminoglycoside phosphotransferase | |
| 4 | View Details | [364..519] | 1.049982 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [520..582] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)