













| General Information: |
|
| Name(s) found: |
IF4F1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 952 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDETAHPTQ SASKQESAAL KQTGDDQQES QQQRGYTNYN NGSNYTQKKP YNSNRPHQQR 60
61 GGKFGPNRYN NRGNYNGGGS FRGGHMGANS SNVPWTGYYN NYPVYYQPQQ MAAAGSAPAN 120
121 PIPVEEKSPV PTKIEITTKS GEHLDLKEQH KAKLQSQERS TVSPQPESKL KETSDSTSTS 180
181 TPTPTPSTND SKASSEENIS EAEKTRRNFI EQVKLRKAAL EKKRKEQLEG SSGNNNIPMK 240
241 TTPENVEEKG SDKPEVTEKT KPAEEKSAEP EVKQETPAEE GEQGEKGQIK EESTPKVLTF 300
301 AERLKLKKQQ KEREEKTEGK ENKEVPVQEE TKSAIESAPV PPSEQVKEET EVAETEQSNI 360
361 DESATTPAIP TKSDEAEAEV EAEAGDAGTK IGLEAEIETT TDETDDGTNT VSHILNVLKD 420
421 ATPIEDVFSF NYPEGIEGPD IKYKKEHVKY TYGPTFLLQF KDKLNVKADA EWVQSTASKI 480
481 VIPPGMGRGN RSRDSGRFGN NSSRGHDFRN TSVRNMDDRA NSRTSSKRRS KRMNDDRRSN 540
541 RSYTSRRDRE RGSYRNEEKR EDDKPKEEVA PLVPSANRWV PKFKSKKTEK KLAPDGKTEL 600
601 LDKDEVERKM KSLLNKLTLE MFDAISSEIL AIANISVWET NGETLKAVIE QIFLKACDEP 660
661 HWSSMYAQLC GKVVKELNPD ITDETNEGKT GPKLVLHYLV ARCHAEFDKG WTDKLPTNED 720
721 GTPLEPEMMS EEYYAAASAK RRGLGLVRFI GFLYRLNLLT GKMMFECFRR LMKDLTDSPS 780
781 EETLESVVEL LNTVGEQFET DSFRTGQATL EGSQLLDSLF GILDNIIQTA KISSRIKFKL 840
841 IDIKELRHDK NWNSDKKDNG PKTIQQIHEE EERQRQLKNN SRSNSRRTNN SSNRHSFRRD 900
901 APPASKDSFI TTRTYSQRNS QRAPPPKEEP AAPTSTATNM FSALMGESDD EE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
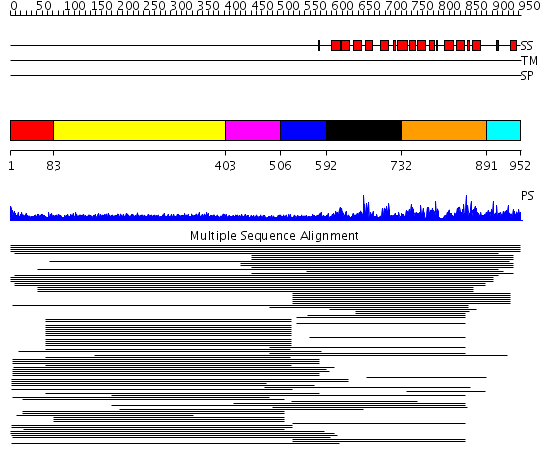
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 12.14 | No description for 1l8mA was found. | |
| 2 | View Details | [83..402] | 10.91 | Hybrid domain of integrin beta; Integrin beta A domain; Integrin beta tail domain; Integrin beta EGF-like domains | |
| 3 | View Details | [403..505] | 10.91 | Hybrid domain of integrin beta; Integrin beta A domain; Integrin beta tail domain; Integrin beta EGF-like domains | |
| 4 | View Details | [506..591] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [592..731] | 310.165202 | Eukaryotic initiation factor eIF4G | |
| 6 | View Details | [732..890] | 310.165202 | Eukaryotic initiation factor eIF4G | |
| 7 | View Details | [891..952] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)