













| General Information: |
|
| Name(s) found: |
EI2BE_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 712 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGKKGQKKS GLGNHGKNSD MDVEDRLQAV VLTDSYETRF MPLTAVKPRC LLPLANVPLI 60
61 EYTLEFLAKA GVHEVFLICS SHANQINDYI ENSKWNLPWS PFKITTIMSP EARCTGDVMR 120
121 DLDNRGIITG DFILVSGDVL TNIDFSKMLE FHKKMHLQDK DHISTMCLSK ASTYPKTRTI 180
181 EPAAFVLDKS TSRCIYYQDL PLPSSREKTS IQIDPELLDN VDEFVIRNDL IDCRIDICTS 240
241 HVPLIFQENF DYQSLRTDFV KGVISSDILG KHIYAYLTDE YAVRVESWQT YDTISQDFLG 300
301 RWCYPLVLDS NIQDDQTYSY ESRHIYKEKD VVLAQSCKIG KCTAIGSGTK IGEGTKIENS 360
361 VIGRNCQIGE NIRIKNSFIW DDCIIGNNSI IDHSLIASNA TLGSNVRLND GCIIGFNVKI 420
421 DDNMDLDRNT KISASPLKNA GSRMYDNESN EQFDQDLDDQ TLAVSIVGDK GVGYIYESEV 480
481 SDDEDSSTEA CKEINTLSNQ LDELYLSDDS ISSATKKTKK RRTMSVNSIY TDREEIDSEF 540
541 EDEDFEKEGI ATVERAMENN HDLDTALLEL NTLRMSMNVT YHEVRIATIT ALLRRVYHFI 600
601 ATQTLGPKDA VVKVFNQWGL LFKRQAFDEE EYIDLMNIIM EKIVEQSFDK PDLILFSALV 660
661 SLYDNDIIEE DVIYKWWDNV STDPRYDEVK KLTVKWVEWL QNADEESSSE EE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
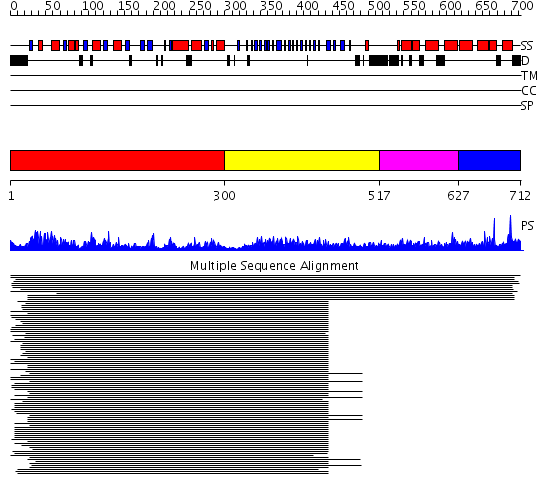
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 116.917458 | N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain; N-acetylglucosamine 1-phosphate uridyltransferase GlmU, N-terminal domain | |
| 2 | View Details | [300..516] | 116.917458 | N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain; N-acetylglucosamine 1-phosphate uridyltransferase GlmU, N-terminal domain | |
| 3 | View Details | [517..626] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [627..712] | 27.60206 | eIF4-gamma/eIF5/eIF2-epsilon No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)