













| General Information: |
|
| Name(s) found: |
DHE2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1092 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLFDNKNRGA LNSLNTPDIA SLSISSMSDY HVFDFPGKDL QREEVIDLLD QQGFIPDDLI 60
61 EQEVDWFYNS LGIDDLFFSR ESPQLISNII HSLYASKLDF FAKSKFNGIQ PRLFSIKNKI 120
121 ITNDNHAIFM ESNTGVSISD SQQKNFKFAS DAVGNDTLEH GKDTIKKNRI EMDDSCPPYE 180
181 LDSEIDDLFL DNKSQKNCRL VSFWAPESEL KLTFVYESVY PNDDPAGVDI SSQDLLKGDI 240
241 ESISDKTMYK VSSNENKKLY GLLLKLVKER EGPVIKTTRS VENKDEIRLL VAYKRFTTKR 300
301 YYSALNSLFH YYKLKPSKFY LESFNVKDDD IIIFSVYLNE NQQLEDVLLH DVEAALKQVE 360
361 REASLLYAIP NNSFHEVYQR RQFSPKEAIY AHIGAIFINH FVNRLGSDYQ NLLSQITIKR 420
421 NDTTLLEIVE NLKRKLRNET LTQQTIINIM SKHYTIISKL YKNFAQIHYY HNSTKDMEKT 480
481 LSFQRLEKVE PFKNDQEFEA YLNKFIPNDS PDLLILKTLN IFNKSILKTN FFITRKVAIS 540
541 FRLDPSLVMT KFEYPETPYG IFFVVGNTFK GFHIRFRDIA RGGIRIVCSR NQDIYDLNSK 600
601 NVIDENYQLA STQQRKNKDI PEGGSKGVIL LNPGLVEHDQ TFVAFSQYVD AMIDILINDP 660
661 LKENYVNLLP KEEILFFGPD EGTAGFVDWA TNHARVRNCP WWKSFLTGKS PSLGGIPHDE 720
721 YGMTSLGVRA YVNKIYETLN LTNSTVYKFQ TGGPDGDLGS NEILLSSPNE CYLAILDGSG 780
781 VLCDPKGLDK DELCRLAHER KMISDFDTSK LSNNGFFVSV DAMDIMLPNG TIVANGTTFR 840
841 NTFHTQIFKF VDHVDIFVPC GGRPNSITLN NLHYFVDEKT GKCKIPYIVE GANLFITQPA 900
901 KNALEEHGCI LFKDASANKG GVTSSSMEVL ASLALNDNDF VHKFIGDVSG ERSALYKSYV 960
961 VEVQSRIQKN AELEFGQLWN LNQLNGTHIS EISNQLSFTI NKLNDDLVAS QELWLNDLKL1020
1021 RNYLLLDKII PKILIDVAGP QSVLENIPES YLKVLLSSYL SSTFVYQNGI DVNIGKFLEF1080
1081 IGGLKREAEA SA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
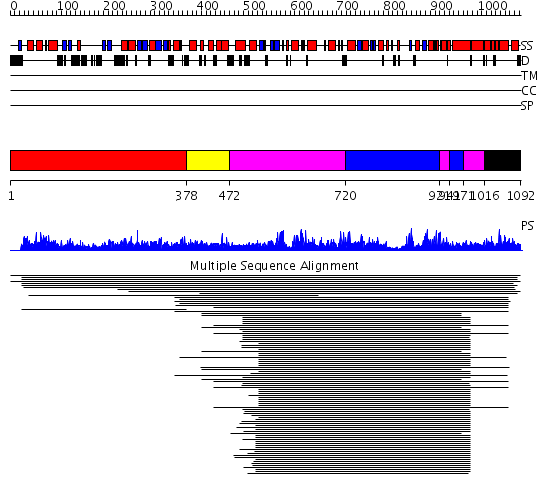
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..377] | 1.016878 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [378..471] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [472..719] [921..940] [971..1015] |
117.0 | Glutamate dehydrogenase | |
| 4 | View Details | [720..920] [941..970] |
117.0 | Glutamate dehydrogenase | |
| 5 | View Details | [1016..1092] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)