













| General Information: |
|
| Name(s) found: |
CBF3B_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 608 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFNRTTQLKS KHPCSVCTRR KVKCDRMIPC GNCRKRGQDS ECMKSTKLIT ASSSKEYLPD 60
61 LLLFWQNYEY WITNIGLYKT KQRDLTRTPA NLDTDTEECM FWMNYLQKDQ SFQLMNFAME 120
121 NLGALYFGSI GDISELYLRV EQYWDRRADK NHSVDGKYWD ALIWSVFTMC IYYMPVEKLA 180
181 EIFSVYPLHE YLGSNKRLNW EDGMQLVMCQ NFARCSLFQL KQCDFMAHPD IRLVQAYLIL 240
241 ATTTFPYDEP LLANSLLTQC IHTFKNFHVD DFRPLLNDDP VESIAKVTLG RIFYRLCGCD 300
301 YLQSGPRKPI ALHTEVSSLL QHAAYLQDLP NVDVYREENS TEVLYWKIIS LDRDLDQYLN 360
361 KSSKPPLKTL DAIRRELDIF QYKVDSLEED FRSNNSRFQK FIALFQISTV SWKLFKMYLI 420
421 YYDTADSLLK VIHYSKVIIS LIVNNFHAKS EFFNRHPMVM QTITRVVSFI SFYQIFVESA 480
481 AVKQLLVDLT ELTANLPTIF GSKLDKLVYL TERLSKLKLL WDKVQLLDSG DSFYHPVFKI 540
541 LQNDIKIIEL KNDEMFSLIK GLGSLVPLNK LRQESLLEEE DENNTEPSDF RTIVEEFQSE 600
601 YNISDILS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
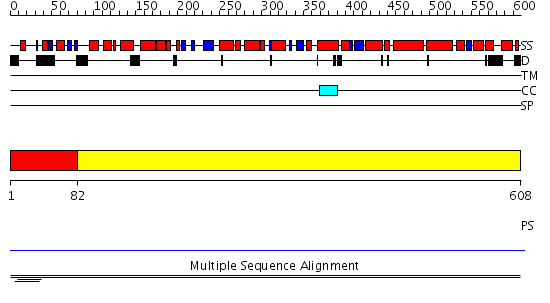
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | 24.071898 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [82..608] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [194..362] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [363..608] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)