













| General Information: |
|
| Name(s) found: |
CA120_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1060 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRIFSGDNKV VDSLASNPGL MSPSNFGGDF GSRLKVNVTS KKKLNDSSPT SPMESSPVSP 60
61 ELVPILTLLN AHTHRRYHEG VFLILQDLNN NGTHAARKWK DVYGVLLGTQ LALWDAKELA 120
121 EFTDPSCPVS EKKLKEVASK PTYINLTDAT LRTLDNSDNI VMECGKNLTN ALVVSTTLKN 180
181 RYFLQFGNKE SFNAWNSAIR LCLYECSSLQ EAYTGAFISS RGAKLGDIRI LLTNRKYDYK 240
241 DWVSVRFGAG MPWKRCYAVI SQSSSKKKGH FGEINLYEND KKVKKNHAMA TIVEAKALYA 300
301 VYPSSPKLID SSTIIKVVGS VKFEKKESAQ EKDVFIMPEK HQAVPSYDTI IRFLIPAMDT 360
361 FKLYGRPEKL LSSKNDPHSL LFGLPVLPHI YYLEVEDLLP LTNSVSSLHW SNNEWKEHIS 420
421 DILQRKIAQG YCGCNSTSNI TSPLPSPFLG SADLFERADG VLSPKLSYGS KSSSNNSSKN 480
481 SLPKRERVKL SSSSEQDLNN SDSPSIKRKS PPLVISESPH KVHTPTDASF RTRVTEGSPY 540
541 AKQRHPKPFA SSVNDSPSDR AKSRTVPYNN NDRKATTPEK FERGETSCGK NVDESLEKVR 600
601 NMKLEIPESN FDKFMTDKNL LSVDSKCSNE KKLSVESDLS AIYEKYSNGP FGHTEGLNGS 660
661 SDETYLRFQR ASVHSESNYN SRKSFTPSDF SNGNEEEHAV LQELNSLTQR INELGMESIN 720
721 SNSDSDRING SYSQVDFGNN NDEDDMNLFD PDFMAQDQLR AEERDYNKDD RTPLAKVPAA 780
781 FQSTGLGITP DDDIERQYIT EHRSRHEVPK RSPEKPSNPL EIGNPYAKPG TRLNTTHTHS 840
841 KTDRSITPQR GQPVPSGQQI SSYVQPANIN SPNKMYGANN SAMGSPRNPK TRAPPGPYNQ 900
901 GWNNRPSPSN IYQRPHPSDT QPQAYHLPGN PYSTGNRPNM QAQYHPQQVP MPILQQPNRP 960
961 YQPYAMNTHM GSPGGYAGAA PPFQPANVNY NTRPQQPWPT PNSPSAHYRP PPNLNQPQNG1020
1021 SAGYYRPPAP QLQNSQARPQ KKDGFSQFMP SATTKNPYAQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
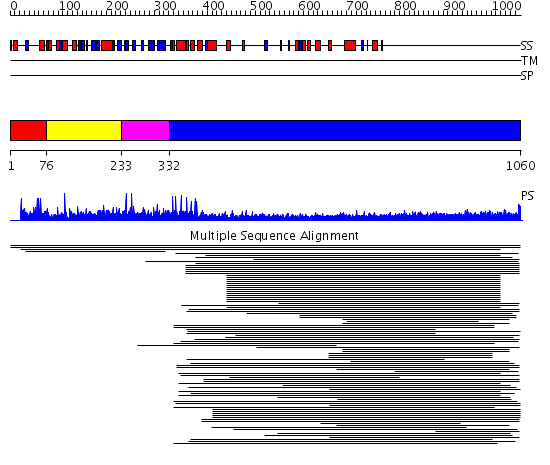
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [76..232] | 8.585027 | PH domain No confident structure predictions are available. | |
| 3 | View Details | [233..331] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [332..1060] | 37.355997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [792..865] | 1.250991 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [866..1060] | 5.247997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)