













| General Information: |
|
| Name(s) found: |
BIR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 954 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGQIDKMEK RYSMTKLENR LRTFQDGVAL EKKKLKWSFK VIPYQAMAKL GFYFDPVIDP 60
61 KTSKLKKDSV RCCYCHRQTY NVRDCRSKRK DVLETLSNIM RQHLTVTDNK QVCLLIYLRN 120
121 KLLTDYSFHM GVSDWKNDKY FSNPDDENVI NLRKFTFQDN WPHSGSQNEH PLGIEKMVNA 180
181 GLMRYDSSIE GLGDPSMDKT LMNDTCYCIY CKQLLQGWSI NDDPMSRHYK VSQNGNCYFF 240
241 QTRNRFERIK NDNDSITKNC EVSPTLGENG KREVINTKTA SQRQCPLFES PPSSTGPQLD 300
301 DYNEKTDISV IQHNISVLDG AQGENVKRNS VEEKEQINME NGSTTLEEGN INRDVLADKK 360
361 EVISTPTAKE IKRPNVQLTQ SSSPIKKKRK FKRISPRKIF DEEDSEHSLN NNSANGDNKD 420
421 KDLVIDFTSH IIKNRDVGRK NAILDDSTDE FSFSNQGHNT FDIPIPTSSH LLKGIDSDND 480
481 NVIREDDTGI NTDTKGASSK HEKFSVNSEE DLNFSEVKLT GRDSSTNILI RTQIVDQNLG 540
541 DIDRDKVPNG GSPEVPKTHE LIRDNSEKRE AQNGEFRHQK DSTVRQSPDI LHSNKSGDNS 600
601 SNITAIPKEE QRRGNSKTSS IPADIHPKPR KNLQEPRSLS ISGKVVPTER KLDNINIDLN 660
661 FSASDFSPSS QSEQSSKSSS VISTPVASPK INLTRSLHAV KELSGLKKET DDGKYFTNKQ 720
721 ETIKILEDVS VKNETPNNEM LLFETGTPIA SQENKSRKLF DEEFSGKELD IPIDSSTVEI 780
781 KKVIKPEFEP VPSVARNLVS GTSSYPRNSR LEEQRKETST SLADNSKKGS SFNEGNNEKE 840
841 PNAAEWFKID ENRHLVKNYF HDLLKYINNN DATLANDKDG DLAFLIKQMP AEELDMTFNN 900
901 WVNLKVQSIK REFIDDCDKK LDILRRDYYT ATNFIETLED DNQLIDIAKK MGIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
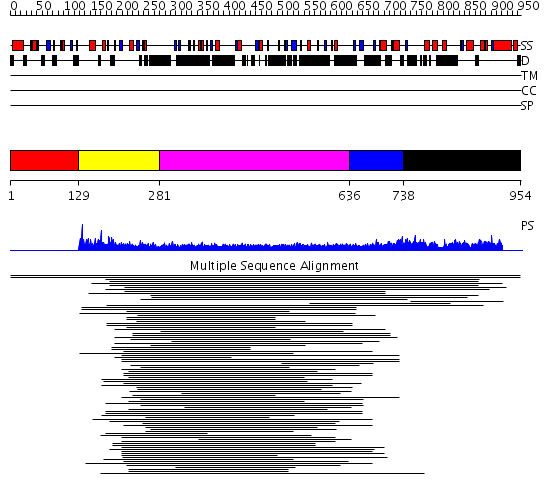
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 15.259637 | Inhibitor of Apoptosis domain No confident structure predictions are available. | |
| 2 | View Details | [129..280] | 14.83 | Anti-apoptotic protein survivin | |
| 3 | View Details | [281..635] | 6.164997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [636..737] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [738..954] | 1.018964 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)