













| General Information: |
|
| Name(s) found: |
SAC9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1630 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to cold
[NAS]
phosphoinositide-mediated signaling [TAS] response to osmotic stress [IMP] response to high light intensity [NAS] phosphatidylinositol metabolic process [IMP] |
| Molecular Function: |
inositol or phosphatidylinositol phosphatase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLHPPGGSK KTSVVVVTLD TGEVYVIASL LSKADTQVIY IDPTTGILRY NGKPGLDNFK 60
61 SEREALDYIT NGSRGGVRSS VYARAILGYA VLGSFGMLLV ATRLNPSIPD LPGGGCVYTV 120
121 AESQWVKIPL YNPQPQGKGE TKNIQELTEL DIDGKHYFCD TRDITRPFPS RMPLQSPDDE 180
181 FVWNRWLSVP FKNIGLPEHC VILLQGFAEY RPFGSSGQLE GIVALMARRS RLHPGTRYLA 240
241 RGINSCSGTG NEVECEQLVW IPKRNGQSIA FNSYIWRRGT IPIWWGAELK MTAAEAEIYV 300
301 ADRDPYKGST EYYQRLSKRY DTRNLDAPVG ENQKKKAFVP IVCVNLLRSG EGKSECILVQ 360
361 HFEESMNFIK SSGKLPYTRV HLINYDWHAS VKLKGEQQTI EGLWMYLKSP TMAIGISEGD 420
421 YLPSRQRLKD CRGEVICIDD IEGAFCLRSH QNGVIRFNCA DSLDRTNAAS FFGGLQVFVE 480
481 QCRRLGISLD TDLGYGHNSV NNQGGYNAPL PPGWEKRADA VTGKSYYIDH NTKTTTWSHP 540
541 CPDKPWKRLD MRFEEFKRST ILSPVSELAD LFLQQGDIHA TLYTGSKAMH SQILNIFSEE 600
601 SGAFKQFSAA QKNMKITLQR RYKNAMVDSS RQKQLEMFLG MRLFKHLPSI PVQPLHVLSR 660
661 PSGFFLKPVP NMSESSNDGS SLLSIKRKDI TWLCPQAADI VELFIYLSEP CHVCQLLLTI 720
721 SHGADDLTCP STVDVRTGRH IEDLKLVVEG ASIPRCANGT NLLVPLPGPI SSEDMAVTGA 780
781 GARLHEKDTS SLSLLYDFEE LEGQLDFLTR VVAVTFYPAG AVRIPMTLGQ IEVLGISLPW 840
841 KGMFTCERTG GRLAELARKP DEDGSPFSSC SDLNPFAATT SLQAETVSTP VQQKDPFPSN 900
901 LLDLLTGEDS SSDPFPQPVV ECIASGGNDM LDFLDEAVVE YRGSDTVPDG SVPQNKRPKD 960
961 SGAHLYLNCL KSLAGPNMAK KLEFVEAMKL EIERLRLNIS AAERDRALLS IGIDPATINP1020
1021 NSSYDELYIG RLCKIANALA VMGQASLEDK IIASIGLEKL ENNVIDFWNI TRIGEGCDGG1080
1081 MCQVRAEVNK SPVGSSTKSS RGESGSVFLC FQCMKKACKF CCAGKGALLL SKSYSRDTAN1140
1141 GGGSLADVSA TSIGSDHYIC KKCCSSIVLE ALIVDYVRVM VSLRRSGRVD NAGREALNEV1200
1201 FGSNITNHLA VRGQPSPNRE DFNFLRQILG KEESLSEFPF ASFLHKVETA TDSAPFFSLL1260
1261 TPLNLASSNA YWKAPPSADS VEAAIVLNTL SDVSSVILLV SPCGYSDADA PTVQIWASSD1320
1321 INKEARTLMG KWDVQSFIRS SPELSGSEKS GRAPRHIKFA FKNPVRCRII WITLRLPRLG1380
1381 SSSSVSLDKN INLLSLDENP FAPIPRRASF GATIENDPCI HAKHILVTGN TVRDKTLQSV1440
1441 ESMSVRNWLD RAPRLNRFLI PLETERPMEN DLVLELYLQP ASPLAAGFRL DAFSAIKPRV1500
1501 THSPSSDVVD IWDPTSVIME DRHVSPAILY IQVSVLQEQY KMVTIAEYRL PEARDGTKLY1560
1561 FDFPKQIQAQ RVSFKLLGDV AAFTDEPAEA VDLSSRASPF AAGLSLANRI KLYYYADPYE1620
1621 VGKWTSLSSV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
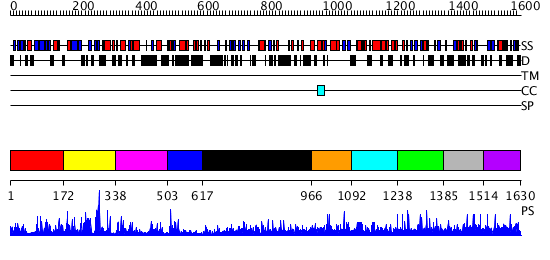
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 5.183992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [172..337] | 1.193981 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [338..502] | 1.279993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [503..616] | 12.0 | Mitotic rotamase PIN1; Mitotic rotamase PIN1, domain 2 | |
| 5 | View Details | [617..965] | 35.0 | Synaptojanin, IPP5C domain | |
| 6 | View Details | [966..1091] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1092..1237] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1238..1384] | 1.005956 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1385..1513] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1514..1630] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|