













| General Information: |
|
| Name(s) found: |
PLPD2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 567 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast envelope [IDA] |
| Biological Process: |
cell redox homeostasis
[IEA]
|
| Molecular Function: |
dihydrolipoyl dehydrogenase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSVLSLSFS QASLPLANRT LCSSNAAPST PRNLRFCGLR REAFCFSPSK QLTSCRFHIQ 60
61 SRRIEVSAAA SSSAGNGAPS KSFDYDLIII GAGVGGHGAA LHAVEKGLKT AIIEGDVVGG 120
121 TCVNRGCVPS KALLAVSGRM RELQNEHHMK AFGLQVSAAG YDRQGVADHA SNLATKIRNN 180
181 LTNSMKALGV DILTGFGAVL GPQKVKYGDN IITGKDIIIA TGSVPFVPKG IEVDGKTVIT 240
241 SDHALKLESV PDWIAIVGSG YIGLEFSDVY TALGSEVTFI EALDQLMPGF DPEISKLAQR 300
301 VLINTRKIDY HTGVFASKIT PAKDGKPVLI ELIDAKTKEP KDTLEVDAAL IATGRAPFTN 360
361 GLGLENINVT TQRGFIPVDE RMRVIDGNGK LVPHLYCIGD ANGKLMLAHA ASAQGISVVE 420
421 QVTGRDHVLN HLSIPAACFT HPEISMVGLT EPQAREKAEK EGFKVSIAKT SFKANTKALA 480
481 ENEGEGLAKM IYRPDNGEIL GVHIFGLHAA DLIHEASNAI ALGTRIQDIK LAVHAHPTLS 540
541 EVVDELFKAA KVDSPASVTA QSVKVTV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
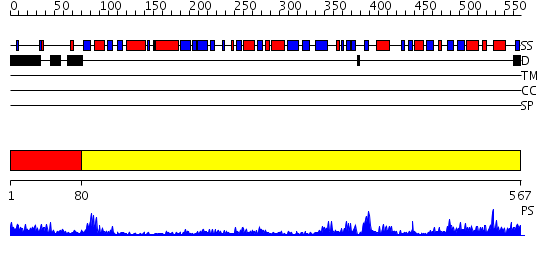
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | 18.69897 | Trimethylamine dehydrogenase, N-terminal domain; Trimethylamine dehydrogenase, C-terminal domain; Trimethylamine dehydrogenase, middle domain | |
| 2 | View Details | [80..567] | 103.0 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|