













| General Information: |
|
| Name(s) found: |
GYS1_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 708 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARDLQNHLL FEVATEVTNR VGGIYSVLKS KAPVTVAQYG DNYTLLGPLN KATYESEVEK 60
61 LDWEDESIFP EELLPIQKTL MSMREKGVNF VYGNWLIEGA PRVILFELDS VRHFLNEWKA 120
121 DLWSLVGIPS PEHDHETNDA ILLGYVVVWF LGEVSKLDSS HAIIGHFHEW LAGVALPLCR 180
181 KKRIDVVTIF TTHATLLGRY LCAAGDVDFY NNLQYFDVDQ EAGKRGIYHR YCIERAAAHT 240
241 ADVFTTVSQI TALEAEHLLK RKPDGILPNG LNVVKFQAVH EFQNLHALKK DKINDFVRGH 300
301 FHGCFDFDLD NTVYFFIAGR YEYKNKGADM FIESLARLNY RLKVSGSKKT VVAFLIMPAK 360
361 TNSFTVEALK SQAIVKSLEN TVNEVTASIG KRIFEHTMRY PHNGLESELP TNLDELLKSS 420
421 EKVLLKKRVL ALRRPYGELP PVVTHNMCDD ANDPILNQIR HVRLFNDSSD RVKVIFHPEF 480
481 LNANNPILGL DYDEFVRGCH LGVFPSYYEP WGYTPAECTV MGVPSITTNV SGFGAYMEDL 540
541 IETDQAKDYG IYIVDRRFKS PDESVEQLAD YMEEFVNKTR RQRINQRNRT ERLSDLLDWK 600
601 RMGLEYVKAR QLGLRRAYPE QFKQLVGETI SDANMNTLAG GKKFKIARPL SVPGSPKVRS 660
661 NSTVYMTPGD LGTLQDANNA DDYFNLSTNG AIDNDDDDND TSAYYEDN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
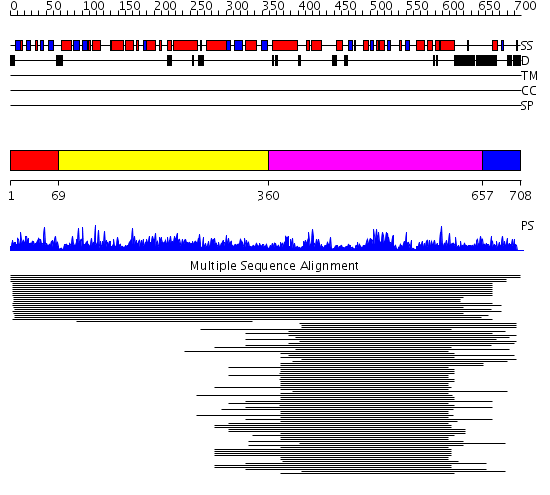
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [69..359] | 1.421879 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [360..656] | 277.951997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [657..708] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)