













| General Information: |
|
| Name(s) found: |
SNU66_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKTENLSIE ETNEIREKLG MKPIPVFQEK NTDHKESLSI EETNELRASL GLKLIPPQQN 60
61 FNSSPPNVHN TSKIDELREK ITKFQKKANA PLRMAHLLEE TNVNDDSSWL ENMDAIPSSH 120
121 ESKRSSTLPR KGATKEDENI DLHNVQVSYN IEALSPKKDT ILTLKESSIF DDTDSTEVLE 180
181 NVKAAEENAD REKLRLRQMN KDRRQKKKIL NVSSLDIEEE EEGEKHSITT THLIIGAEQG 240
241 VMKAPNTISA KPPTGKVKVN FDSANNMSDE DGGDFKPLKI KKRKIKDPRS TKARKSKITD 300
301 KMEIVKLVDE DESLSWMEEE QPVTIINPRT SSNNELKGPE DLAREIEKAR DEEKRRTESI 360
361 LKMREISNSI VVDEKVTFLN TLDTSLSERS ATENKVKVHG EGEKNIGDVT NGHTKEGSGN 420
421 NTLTEAVNNE PNYEGDAENA PNFFSGLAST LGYLRKKSVF TTGDVDLKPG KDVNNSESLR 480
481 RDVRNKEHTG TGTYTKDKLH GLEQFTSSDS SNANTHSKRQ DHYDPDIKLV YRDEKGNRLT 540
541 TKEAYKKLSQ KFHGTKSNKK KRAKMKSRIE ARKNTPENGS LFEFDDN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
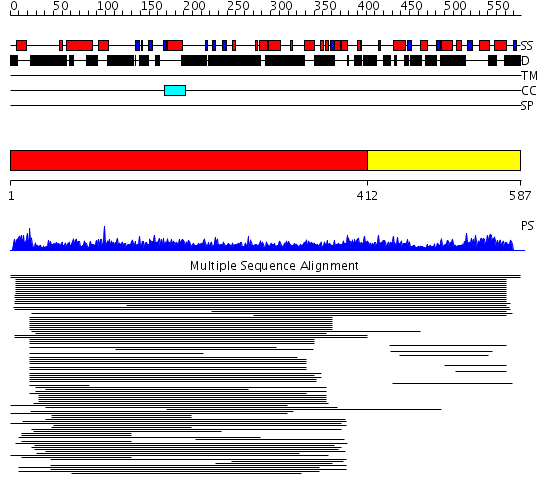
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..411] | 2.09691 | Heavy meromyosin subfragment | |
| 2 | View Details | [412..587] | 2.020986 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [271..438] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [439..587] | 0.96 | Large T antigen, the N-terminal J domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)