













| General Information: |
|
| Name(s) found: |
mutM /
EG10329
[EcoGene]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 269 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA repair
[IEA]
base-excision repair [IEA] metabolic process [IEA] nucleotide-excision repair [IEA] response to DNA damage stimulus [IDA |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA] zinc ion binding [IDA metal ion binding [IDA endonuclease activity [UNKNOWN lyase activity [IEA] DNA N-glycosylase activity [UNKNOWN DNA-(apurinic or apyrimidinic site) lyase activity [IDA damaged DNA binding [IDA oxidized purine base lesion DNA N-glycosylase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPELPEVETS RRGIEPHLVG ATILHAVVRN GRLRWPVSEE IYRLSDQPVL SVQRRAKYLL 60
61 LELPEGWIII HLGMSGSLRI LPEELPPEKH DHVDLVMSNG KVLRYTDPRR FGAWLWTKEL 120
121 EGHNVLTHLG PEPLSDDFNG EYLHQKCAKK KTAIKPWLMD NKLVVGVGNI YASESLFAAG 180
181 IHPDRLASSL SLAECELLAR VIKAVLLRSI EQGGTTLKDF LQSDGKPGYF AQELQVYGRK 240
241 GEPCRVCGTP IVATKHAQRA TFYCRQCQK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
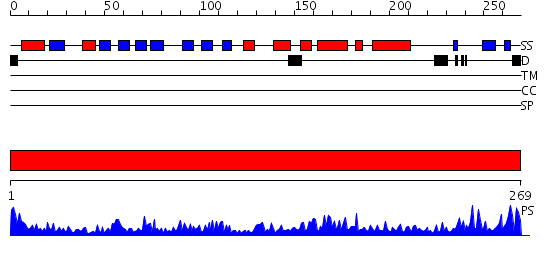
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..269] | 106.0 | DNA repair protein MutM (Fpg) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)