













| General Information: |
|
| Name(s) found: |
araD /
EG10055
[EcoGene]
|
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 231 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
L-lyxose metabolic process
[IMP L-arabinose catabolic process to xylulose 5-phosphate [IEP |
| Molecular Function: |
zinc ion binding
[IEA]
metal ion binding [IEA] L-ribulose-phosphate 4-epimerase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLEDLKRQVL EANLALPKHN LVTLTWGNVS AVDRERGVFV IKPSGVDYSV MTADDMVVVS 60
61 IETGEVVEGT KKPSSDTPTH RLLYQAFPSI GGIVHTHSRH ATIWAQAGQS IPATGTTHAD 120
121 YFYGTIPCTR KMTDAEINGE YEWETGNVIV ETFEKQGIDA AQMPGVLVHS HGPFAWGKNA 180
181 EDAVHNAIVL EEVAYMGIFC RQLAPQLPDM QQTLLDKHYL RKHGAKAYYG Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
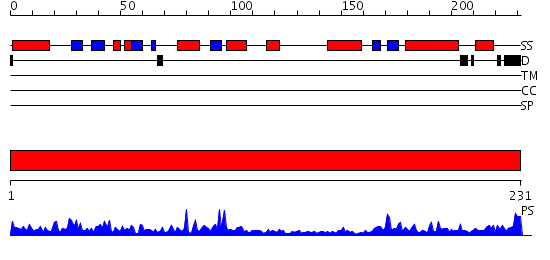
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..231] | 66.0 | L-ribulose-5-phosphate 4-epimerase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)