













| General Information: |
|
| Name(s) found: |
gpsA /
EG20091
[EcoGene]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 339 amino acids |
Gene Ontology: |
|
| Cellular Component: |
glycerol-3-phosphate dehydrogenase complex
[IEA]
cytoplasm [IEA] |
| Biological Process: |
glycerol-3-phosphate metabolic process
[IEA]
carbohydrate metabolic process [IEA] glycerol-3-phosphate biosynthetic process [IEA] glycerol-3-phosphate catabolic process [IEA] glycerophospholipid biosynthetic process [IMP metabolic process [IEA] phospholipid biosynthetic process [IEA] oxidation reduction [IEA] |
| Molecular Function: |
oxidoreductase activity, acting on CH-OH group of donors
[IEA]
coenzyme binding [IEA] oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor [IEA] catalytic activity [IEA] NAD or NADH binding [IEA] glycerol-3-phosphate dehydrogenase [NAD(P)+] activity [IEA] binding [IEA] glyceraldehyde-3-phosphate dehydrogenase activity [IMP glycerol-3-phosphate dehydrogenase (NAD+) activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNQRNASMTV IGAGSYGTAL AITLARNGHE VVLWGHDPEH IATLERDRCN AAFLPDVPFP 60
61 DTLHLESDLA TALAASRNIL VVVPSHVFGE VLRQIKPLMR PDARLVWATK GLEAETGRLL 120
121 QDVAREALGD QIPLAVISGP TFAKELAAGL PTAISLASTD QTFADDLQQL LHCGKSFRVY 180
181 SNPDFIGVQL GGAVKNVIAI GAGMSDGIGF GANARTALIT RGLAEMSRLG AALGADPATF 240
241 MGMAGLGDLV LTCTDNQSRN RRFGMMLGQG MDVQSAQEKI GQVVEGYRNT KEVRELAHRF 300
301 GVEMPITEEI YQVLYCGKNA REAALTLLGR ARKDERSSH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
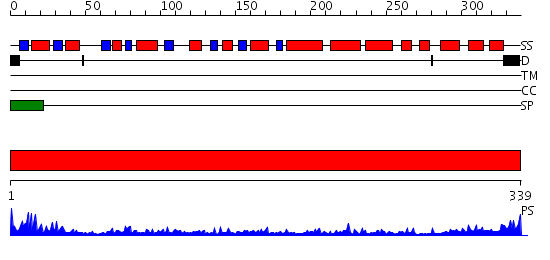
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..339] | 56.39794 | Crystal structure of glycerol-3-phosphate dehydrogenase (TM0378) from THERMOTOGA MARITIMA at 2.00 A resolution |
| Source: Reynolds et al. 2008. Manuscript submitted |

|