













| General Information: |
|
| Name(s) found: |
NADB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 651 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
NAD biosynthetic process
[IGI][IMP][ISS]
|
| Molecular Function: |
oxidoreductase activity
[IEA]
electron carrier activity [IEA] L-aspartate oxidase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAHVSTGNI HNFYLAGQVY RGQAFSWSSA STFMANPFKE PSWSSGVFKA LKAERCGCYS 60
61 RGISPISETS KPIRAVSVSS STKYYDFTVI GSGVAGLRYA LEVAKQGTVA VITKDEPHES 120
121 NTNYAQGGVS AVLCPLDSVE SHMRDTMVAG AHLCDEETVR VVCTEGPERI RELIAMGASF 180
181 DHGEDGNLHL AREGGHSHCR IVHAADMTGR EIERALLEAV LNDPNISVFK HHFAIDLLTS 240
241 QDGLNTVCHG VDTLNIKTNE VVRFISKVTL LASGGAGHIY PSTTNPLVAT GDGMAMAHRA 300
301 QAVISNMEFV QFHPTALADE GLPIKLQTAR ENAFLITEAV RGDGGILYNL GMERFMPVYD 360
361 ERAELAPRDV VARSIDDQLK KRNEKYVLLD ISHKPREKIL AHFPNIASEC LKHGLDITRQ 420
421 PIPVVPAAHY MCGGVRAGLQ GETNVLGLFV AGEVACTGLH GANRLASNSL LEALVFARRA 480
481 VQPSTELMKR TRLDVCASEK WTRPVVATAR LLGDEVIAKI IALTKEVRRE LQEVMWKYVG 540
541 IVRSTIRLTT AERKIAELEA KWETFLFEHG WEQTVVALEA CEMRNLFCCA KLVVSSALAR 600
601 HESRGLHYMT DFPFVEESKR IPTIILPSSP TTASWSSRRL QNISSSSLID C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
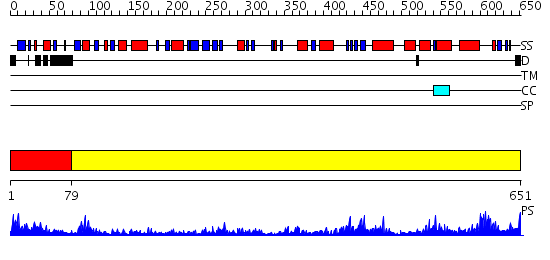
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 17.39794 | No description for 2qy6A was found. | |
| 2 | View Details | [79..651] | 118.0 | L-aspartate oxidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)