













| General Information: |
|
| Name(s) found: |
PMS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 923 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC]
|
| Biological Process: |
fruit development
[IMP]
DNA recombination [IMP] pollen development [IMP] seed development [IMP] mismatch repair [IMP][ISS] |
| Molecular Function: |
ATP binding
[IEA][ISS]
mismatched DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQGDSSPSPT TTSSPLIRPI NRNVIHRICS GQVILDLSSA VKELVENSLD AGATSIEINL 60
61 RDYGEDYFQV IDNGCGISPT NFKVLALKHH TSKLEDFTDL LNLTTYGFRG EALSSLCALG 120
121 NLTVETRTKN EPVATLLTFD HSGLLTAEKK TARQIGTTVT VRKLFSNLPV RSKEFKRNIR 180
181 KEYGKLVSLL NAYALIAKGV RFVCSNTTGK NPKSVVLNTQ GRGSLKDNII TVFGISTFTS 240
241 LQPVSICVSE DCRVEGFLSK PGQGTGRNLA DRQYFFINGR PVDMPKVSKL VNELYKDTSS 300
301 RKYPVTILDF IVPGGACDLN VTPDKRKVFF SDETSVIGSL REGLNEIYSS SNASYIVNRF 360
361 EENSEQPDKA GVSSFQKKSN LLSEGIVLDV SSKTRLGEAI EKENPSLREV EIDNSSPMEK 420
421 FKFEIKACGT KKGEGSLSVH DVTHLDKTPS KGLPQLNVTE KVTDASKDLS SRSSFAQSTL 480
481 NTFVTMGKRK HENISTILSE TPVLRNQTSS YRVEKSKFEV RALASRCLVE GDQLDDMVIS 540
541 KEDMTPSERD SELGNRISPG TQADNVERHE REHEKPIRFE EPTSDNTLTK GDVERVSEDN 600
601 PRCSQPLRSV ATVLDSPAQS TGPKMFSTLE FSFQNLRTRR LERLSRLQST GYVSKCMNTP 660
661 QPKKCFAAAT LELSQPDDEE RKARALAAAT SELERLFRKE DFRRMQVLGQ FNLGFIIAKL 720
721 ERDLFIVDQH AADEKFNFEH LARSTVLNQQ PLLQPLNLEL SPEEEVTVLM HMDIIRENGF 780
781 LLEENPSAPP GKHFRLRAIP YSKNITFGVE DLKDLISTLG DNHGECSVAS SYKTSKTDSI 840
841 CPSRVRAMLA SRACRSSVMI GDPLRKNEMQ KIVEHLADLE SPWNCPHGRP TMRHLVDLTT 900
901 LLTLPDDDNV NDDDDDDATI SLA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
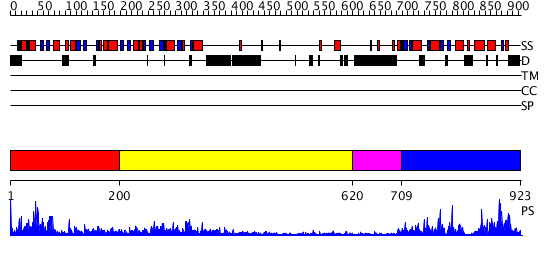
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 70.39794 | DNA mismatch repair protein PMS2 | |
| 2 | View Details | [200..619] | 33.69897 | No description for 2cgeA was found. | |
| 3 | View Details | [620..708] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [709..923] | 18.69897 | Crystal structure of the MutL C-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)