













| General Information: |
|
| Name(s) found: |
POLIB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1034 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleoid
[IDA]
chloroplast [IDA] mitochondrion [IDA] |
| Biological Process: |
DNA replication
[IEA][ISS]
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IEA] |
| Molecular Function: |
nucleic acid binding
[IEA]
3'-5' exonuclease activity [IEA] DNA binding [IEA][ISS] DNA-directed DNA polymerase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVSLRHLSP SSFWVSRRPR VSSSILSFLV PRRRILCTRK VAIIKGNAGY STATDCGGSH 60
61 GFHHSGHQRS SSVEFSGEWK LNLGSKTARM VPPTVKQAGA VSAWREEVNN KLRGRNREYA 120
121 NNQDDAFGNG SYILKGFVPK IDDVHSYGNG QNFDYNLKPG TDITTLGREL NGFMQTNSIR 180
181 GSVVALPSKD IEVGETTDVT LKPLNSDTTL DNASYKKTAT ISKVEKCTNL SQVRANLKKI 240
241 YNRVRVVDNV SSAKETVALL MNQYRNLVHA CDTEVSRIDV KTETPVDHGE MICFSIYCGS 300
301 EADFGDGKSC IWVDVLGENG RDILAEFKPF FEDSSIKKVW HNYSFDNHII RNYGIKLSGF 360
361 HGDTMHMARL WDSSRRISGG YSLEALTSDP KVLGGTETKE EAELFGKISM KKIFGKGKLK 420
421 KDGSEGKLVI IPPVKELQME DREAWISYSA LDSISTLKLY ESMKKQLQAK KWFLDGKLIS 480
481 KKNMFDFYQE YWQPFGELLA KMESEGMLVD RDYLAQIEIV AKAEQEIAVS RFRNWASKHC 540
541 PDAKHMNVGS DTQLRQLFFG GISNSCNDED LPYEKLFKVP NVDKVIEEGK KRATKFRNIK 600
601 LHRISDRPLP TEKFTASGWP SVSGDTLKAL AGKVSAEYDY MEGVLDTCLE ENIGDDDCIS 660
661 LPDEVVETQH VNTSVESDTS AYGTAFDAFG GGESGKEACH AIAALCEVCS IDSLISNFIL 720
721 PLQGSNVSGK DGRVHCSLNI NTETGRLSAR RPNLQNQPAL EKDRYKIRQA FIASPGNSLI 780
781 VADYGQLELR ILAHLASCES MKEAFIAGGD FHSRTAMNMY PHIREAVENG EVLLEWHPQP 840
841 GQEKPPVPLL KDAFASERRK AKMLNFSIAY GKTAIGLSRD WKVSREEAQD TVNLWYNDRQ 900
901 EVRKWQELRK KEAIQKGYVL TLLGRARKFP EYRSRAQKNH IERAAINTPV QGSAADVAMC 960
961 AMLEISNNQR LKELGWKLLL QVHDEVILEG PSESAENAKD IVVNCMSEPF NGKNILSVDL1020
1021 SVDAKCAQNW YAGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
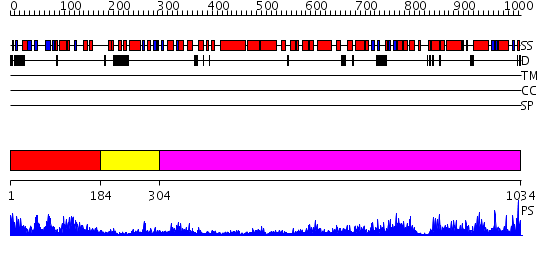
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 21.0 | T5 5'-exonuclease | |
| 2 | View Details | [184..303] | 11.045757 | No description for 2hbjA was found. | |
| 3 | View Details | [304..1034] | 139.0 | 5' to 3' exonuclease domain of DNA polymerase Taq; Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)