













| General Information: |
|
| Name(s) found: |
ATML1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 762 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
epidermal cell differentiation
[IGI]
regulation of transcription, DNA-dependent [ISS] cotyledon development [IGI] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] sequence-specific DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYHPNMFESH HHMFDMTPKN SENDLGITGS HEEDFETKSG AEVTMENPLE EELQDPNQRP 60
61 NKKKRYHRHT QRQIQELESF FKECPHPDDK QRKELSRELS LEPLQVKFWF QNKRTQMKAQ 120
121 HERHENQILK SENDKLRAEN NRYKDALSNA TCPNCGGPAA IGEMSFDEQH LRIENARLRE 180
181 EIDRISAIAA KYVGKPLMAN SSSFPQLSSS HHIPSRSLDL EVGNFGNNNN SHTGFVGEMF 240
241 GSSDILRSVS IPSEADKPMI VELAVAAMEE LVRMAQTGDP LWVSSDNSVE ILNEEEYFRT 300
301 FPRGIGPKPI GLRSEASRES TVVIMNHINL IEILMDVNQW SSVFCGIVSR ALTLEVLSTG 360
361 VAGNYNGALQ VMTAEFQVPS PLVPTRENYF VRYCKQHSDG IWAVVDVSLD SLRPSPITRS 420
421 RRRPSGCLIQ ELQNGYSKVT WVEHIEVDDR SVHNMYKPLV NTGLAFGAKR WVATLDRQCE 480
481 RLASSMASNI PACDLSVITS PEGRKSMLKL AERMVMSFCT GVGASTAHAW TTLSTTGSDD 540
541 VRVMTRKSMD DPGRPPGIVL SAATSFWIPV APKRVFDFLR DENSRSEWDI LSNGGLVQEM 600
601 AHIANGRDPG NSVSLLRVNS GNSGQSNMLI LQESCTDASG SYVIYAPVDI IAMNVVLSGG 660
661 DPDYVALLPS GFAILPDGSA RGGGGSANAS AGAGVEGGGE GNNLEVVTTT GSCGGSLLTV 720
721 AFQILVDSVP TAKLSLGSVA TVNSLIKCTV ERIKAALACD GA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
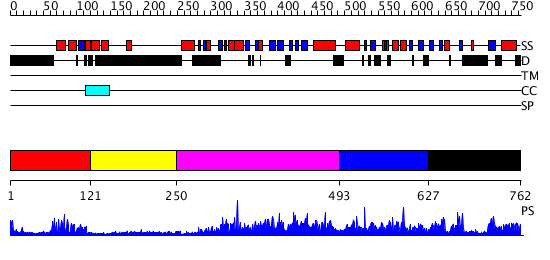
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 17.0 | Homeobox protein hox-b1 | |
| 2 | View Details | [121..249] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [250..492] | 2.73 | No description for 2r55A was found. | |
| 4 | View Details | [493..626] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [627..762] | 2.062966 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)