













| General Information: |
|
| Name(s) found: |
PIF1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 478 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
chlorophyll biosynthetic process
[IMP]
negative gravitropism [IMP][TAS] negative regulation of seed germination [IGI][IMP] regulation of photomorphogenesis [TAS] red light signaling pathway [IDA] heme biosynthetic process [IMP] regulation of seed germination [TAS] gibberellic acid mediated signaling pathway [IMP] regulation of transcription [IMP] negative regulation of photomorphogenesis [IMP] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [ISS][TAS] phytochrome binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHHFVPDFDT DDDYVNNHNS SLNHLPRKSI TTMGEDDDLM ELLWQNGQVV VQNQRLHTKK 60
61 PSSSPPKLLP SMDPQQQPSS DQNLFIQEDE MTSWLHYPLR DDDFCSDLLF SAAPTATATA 120
121 TVSQVTAARP PVSSTNESRP PVRNFMNFSR LRGDFNNGRG GESGPLLSKA VVRESTQVSP 180
181 SATPSAAASE SGLTRRTDGT DSSAVAGGGA YNRKGKAVAM TAPAIEITGT SSSVVSKSEI 240
241 EPEKTNVDDR KRKEREATTT DETESRSEET KQARVSTTST KRSRAAEVHN LSERKRRDRI 300
301 NERMKALQEL IPRCNKSDKA SMLDEAIEYM KSLQLQIQMM SMGCGMMPMM YPGMQQYMPH 360
361 MAMGMGMNQP IPPPSFMPFP NMLAAQRPLP TQTHMAGSGP QYPVHASDPS RVFVPNQQYD 420
421 PTSGQPQYPA GYTDPYQQFR GLHPTQPPQF QNQATSYPSS SRVSSSKESE DHGNHTTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
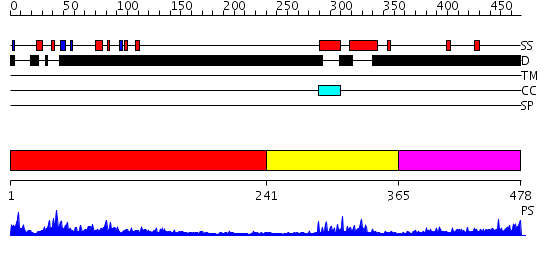
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..240] | 6.291997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [241..364] | 13.39794 | Myc prot-oncogene protein | |
| 3 | View Details | [365..478] | 1.157996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)