













| General Information: |
|
| Name(s) found: |
AT18H_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 927 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
response to starvation
[IEP]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSNSKVNIN NNGDNHNQTK NNGTNGFLPN SLKFISTCIR TASSGVRSAS ASVAASLSSD 60
61 SHELKDQVLW SSFDRLHTSE SSFKNVLLLG YTNGFQVLDI DDSNDVTEFV SRRDDPVTFL 120
121 QMQPLPAKCD GVEGFRSSHP ILLAVADEAK GSGPIVTSRD GSVRNGYEDP LALSPTVVRF 180
181 YSLRSHNYVH VLRFRSTVYM VRCSPRIVAV GLGSQIYCFD ALTLENKFSV LSYPVPQLGN 240
241 QGISGVNVGY GPMAVGARWL AYASNSPLSS SIGRLSPQNV TPPGVSPSTS PNNGNLVARY 300
301 AMESSKHLAA GLLNLGDKGY KTISKYCQDL KHDGPGPSLS SSPGRKVGRV GSHSAESDVV 360
361 GTVIVKDFES RAIIAQFRAH TSPISALCFD PSGTLLVTAS IHGNNINVFR IMPTPTKNGP 420
421 GAQSYDWSSS HVPLYKLHRG MTSAVIQDIC FSSYSQWIAI VSSKSTCHIY VLSPFGGENV 480
481 LEIRNSQFDG PTLAPTLSLP WWSSPSFMTT HFSYPPPASV TLSVVSRIKC NNFFHAASSV 540
541 VGKPTFPSGC LAAVFHQSVP QESQSSSPAL DYLLVYTPSG HVVQYKLIPS LGGDQAESNT 600
601 RNGATSGLTS EEELRVKVEP VQCWDVCRRA DWPEREENIC GLTYGGRKNA ELTVDTSDSE 660
661 DQTKPLEKHH VYLANAEVLI NSGRKPIWQN SEISFYPMYP PDSDGKNLNS HQGGGETEIG 720
721 KVSANEVDIR RKDLLPVYDN FHSVYTSMRN RGFSGERDSD SSSSSDPGQV KEMHPFNGMV 780
781 YPEDEERRGS AHFALTPNQN PHTGIVTFKQ PVVSISSAVK DTDYIDDDAH VLPKNASLPA 840
841 ETKIENSSGI SGDSNVSSNR SDMSMNAADE GEGPIDGSPN FEQFFEEVVS NETVTETEHK 900
901 DAPSDGKLDD DEDDDMLGGV FAFSEEG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
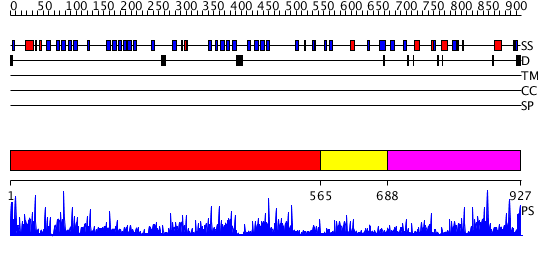
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..564] | 44.221849 | Actin interacting protein 1 | |
| 2 | View Details | [565..687] | 62.09691 | No description for 2ovpB was found. | |
| 3 | View Details | [688..927] | 8.0 | No description for 2vduB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)