













| General Information: |
|
| Name(s) found: |
PDF2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 743 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
epidermal cell differentiation
[IMP]
regulation of transcription, DNA-dependent [ISS] cotyledon development [IGI] |
| Molecular Function: |
DNA binding
[IDA][ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYHPNMFESH HMFDMTPKST SDNDLGITGS REDDFETKSG TEVTTENPSG EELQDPSQRP 60
61 NKKKRYHRHT QRQIQELESF FKECPHPDDK QRKELSRDLN LEPLQVKFWF QNKRTQMKAQ 120
121 SERHENQILK SDNDKLRAEN NRYKEALSNA TCPNCGGPAA IGEMSFDEQH LRIENARLRE 180
181 EIDRISAIAA KYVGKPLGSS FAPLAIHAPS RSLDLEVGNF GNQTGFVGEM YGTGDILRSV 240
241 SIPSETDKPI IVELAVAAME ELVRMAQTGD PLWLSTDNSV EILNEEEYFR TFPRGIGPKP 300
301 LGLRSEASRQ SAVVIMNHIN LVEILMDVNQ WSCVFSGIVS RALTLEVLST GVAGNYNGAL 360
361 QVMTAEFQVP SPLVPTRENY FVRYCKQHSD GSWAVVDVSL DSLRPSTPIL RTRRRPSGCL 420
421 IQELPNGYSK VTWIEHMEVD DRSVHNMYKP LVQSGLAFGA KRWVATLERQ CERLASSMAS 480
481 NIPGDLSVIT SPEGRKSMLK LAERMVMSFC SGVGASTAHA WTTMSTTGSD DVRVMTRKSM 540
541 DDPGRPPGIV LSAATSFWIP VAPKRVFDFL RDENSRKEWD ILSNGGMVQE MAHIANGHEP 600
601 GNCVSLLRVN SGNSSQSNML ILQESCTDAS GSYVIYAPVD IVAMNVVLSG GDPDYVALLP 660
661 SGFAILPDGS VGGGDGNQHQ EMVSTTSSGS CGGSLLTVAF QILVDSVPTA KLSLGSVATV 720
721 NSLIKCTVER IKAAVSCDVG GGA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
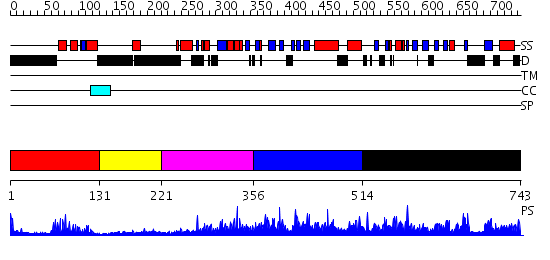
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..130] | 19.522879 | Paired protein | |
| 2 | View Details | [131..220] | 1.21 | No description for 2gd7A was found. | |
| 3 | View Details | [221..355] | 4.522879 | Smad1 | |
| 4 | View Details | [356..513] | 1.7 | No description for 2r55A was found. | |
| 5 | View Details | [514..743] | 1.12 | No description for 2ns9A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)