













| General Information: |
|
| Name(s) found: |
ARP5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 724 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFVSRIRRQ SDYNTYPSSI PIVIDNGASY FRIGWAGETE PRVVFRNIVQ RPRHKATGET 60
61 VTIVGDLDPS MMKYFDCTRS GPRSPFDSNV VYQFEIMEYI LDYAFDRLGA NGSGIDHPIL 120
121 ITECACNPVQ SRSKMAELLF ETYGVPAVAF GVDAAFSYKY NQLHGICKKD GIVLCPGFTT 180
181 THSIPFVDGE PIYKGSSRTN IGGYHVTDYL KQLLSLKYPF HSSRFTWEKA EDLKLEHCYI 240
241 APDYASEIRL FQEGRKEAEE KTSYWQLPWI PPPTEVPPSE EEIARKAAIR EKQGQRLREM 300
301 AEAKRVSKIN DMENQLISLR FLLKQVDQVE EDDIPTFLSD TGYASRQELE STITKVTQSL 360
361 RKARGEPKNE PAEYEENPDS LNNEKYPLMN VPDDILTPEQ LKDKKRQMFL KTTAEGRLRA 420
421 RQKRNEEELE KEKRNQLEEE RRRENPESYL EELQAQYKEV LERVEQKKRL KTNGSSNGNN 480
481 KSGGIGRGER LSAAQRERMR LLTTAAFDRG KGEDTFGSRD EDWQLYKLMS KDNDDDDEQP 540
541 DSDEAELARL SSRLQEIDPT FVQKVEGELS QTSGEVPRVR PLTEEDYKIV IGIERFRCPE 600
601 ILFHPNLIGI DQVGLDEMAG TSIRRLPHDE KELEERLTSS ILMTGGCSLL PGMNERLECG 660
661 IRMIRPCGSP INVVRAMDPV LDAWRGASAF AANLNFLGNA FTKMDYDEKG EDWLRNYQIR 720
721 YNYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
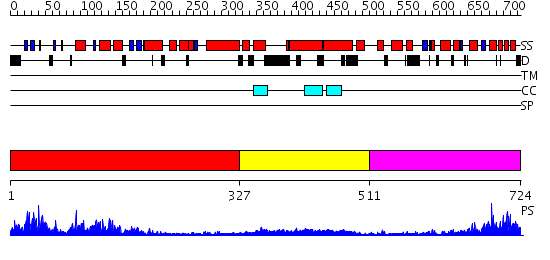
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..326] | 102.0 | Actin | |
| 2 | View Details | [327..510] | 14.69897 | Heavy meromyosin subfragment | |
| 3 | View Details | [511..724] | 6.57 | Actin-related protein 3, Arp3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)