













| General Information: |
|
| Name(s) found: |
gi|22655342
[NCBI NR]
gi|22329713 [NCBI NR] gi|18086338 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 756 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA]
response to stress [IEA] |
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTENGVVTGG GHTVMVGVKF DESSNELLDW ALVKVAEPGD TVIALHVLGN EIVDRADNSS 60
61 LVSIVKNFDS VLEVYEGFCK LKQLELKLKL SRGSSTRKIL VKEAKMCSAS KVVVGISRRF 120
121 HTIHSSVSVA KYLARKVAKD CWVLAVDNGK VMFQKDGSSS IIHHSKGKSD ARRNTLSSFF 180
181 QMPVTLRKNT KVVNHSEVEE EEAEEDHSNG QSLRRSLVYA CLGNCSVRDM NSLPTPGNLS 240
241 RSSSCNGDQD DNADLHKAMA LVPAKFPEDL TPFITMLVKE LPEFRPGWPL LCRVASSDVL 300
301 ASAPRSSSFR KIPVVQWVLK LPARTNSVVG SSDTKQIGFD SSESEENDKL SSSNVERQAI 360
361 VPDESMIVKC SLDHSSGRFP ENVEGLQARI STSCQFFTYK ELVSVTSNFC ADNFIGKGGS 420
421 SRVFRGYLPN GREVAVKILK RTECVLKDFV AEIDIITTLH HKNVISLLGY CFENNNLLLV 480
481 YNYLSRGSLE ENLHGNKKDL VAFRWNERYK VAVGIAEALD YLHNDAPQPV IHRDVKSSNI 540
541 LLSDDFEPQL SDFGLAKWAS ESTTQIICSD VAGTFGYLAP EYFMYGKMNN KIDVYAYGVV 600
601 LLELLSGRKP VNSESPKAQD SLVMWAKPIL DDKEYSQLLD SSLQDDNNSD QMEKMALAAT 660
661 LCIRHNPQTR PTMGMVLELL KGDVEMLKWA KLQVSNPLED SMLLKDEKLR RSNLQSHLNL 720
721 AFLDMEDDSL SMGSMEQGIS VEEYLKGRTS RSSSFN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
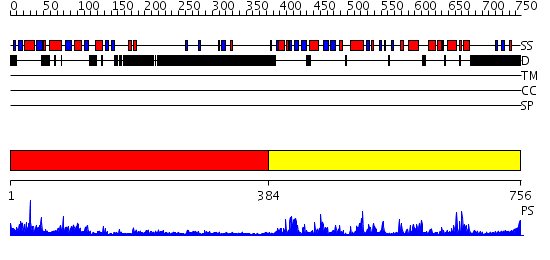
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..383] | 3.522879 | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan | |
| 2 | View Details | [384..756] | 72.69897 | No description for 2qkwB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)