













| General Information: |
|
| Name(s) found: |
VCL1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 858 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plant-type vacuole membrane
[IDA]
|
| Biological Process: |
vacuole organization
[TAS]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANVSVAAEW QLLYDRYYRK PEIYQMKWKH VDLSRNKVAC ASFGGPIAVI RDDSKIVQLY 60
61 AESALRKLRI FNSAGILLSE TVWKHPGGRL IGMSWSDDQT LICIVQDGTI YRYNIHAELI 120
121 EPNMSMGQEC FEQNVVECVF WGNGVVCLTE GGQLICIFDF KTMKPSKLPD VPGLAEDDLL 180
181 QPICLTVREP KYTMSGIAEV LVAVGDDIFV VEEDMVQTIR FDEPSVDDSE MQNDDSGNLI 240
241 GVVQKMIVSP NGKFLTLFTH DGRIVVVDME TKQIAIDYSC ESALPPQQMA WCGMDSVLLY 300
301 WDEDLMMVGP VGDPVHYFYD EPIILIPECD GVRILSNTNL EFLQRVPDST ESIFKIGSTS 360
361 PAALLYDALD HFDRRSAKAD ENLRLIRSSL SEAVESCIDA AGHEFDVTRQ RALLRAASYG 420
421 QAFCSNFQRE RVQETCRTLR VLNAVRDPAI GIPLSIQQYK LLTPVVLISR LINANCHLLA 480
481 LRISEYLDMN KEVVIMHWAC AKITASPSTP DSHLLEILLD KLQLCKGISY AAVATHADNC 540
541 GRRKLAAMLV EHEPRSTKQV PLLLSIGEED TALVKATESG DTDLVYLVIF HIWQKRPPLE 600
601 FFAMIQGRVL ARDLFVAYAR CHKHEFLKDF FLSTGQIHEV AFLLWKESWD MGKNPMASKG 660
661 SPLHGPRIKL IEKARNLFSQ TKEHTFESKA AEEHAKLLKI QHELEASTKQ AIFVDSSIND 720
721 TIRTCIVLGN NRAAIKVKTE FKVSDKRWYW LKAFALATIK DWAALEKFSK EKRPPMGFRP 780
781 FVEACIDADE KAEALKYIPK LSDLVERGEA YARIGMAKEA ADCAAQANDG GELLERFRKT 840
841 FVQNAIFDTL LMPFQGAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
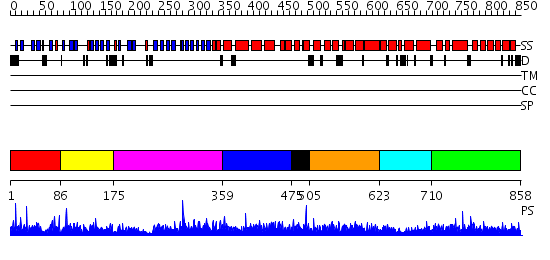
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 1.03499 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [86..174] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [175..358] | 1.048988 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [359..474] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [475..504] | 1.058995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [505..622] | 2.070996 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [623..709] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [710..858] | 5.066992 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)