













| General Information: |
|
| Name(s) found: |
PTBP3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 432 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
mRNA processing
[IEA]
|
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] RNA binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAESSKVVHV RNVGHEISEN DLLQLFQPFG VITKLVMLRA KNQALLQMQD VSSAVSALQF 60
61 FTNVQPTIRG RNVYVQFSSH QELTTIEQNI HGREDEPNRI LLVTIHHMLY PITVDVLHQV 120
121 FSPYGFVEKL VTFQKSAGFQ ALIQYQVQQC AASARTALQG RNIYDGCCQL DIQFSNLEEL 180
181 QVNYNNDRSR DYTNPNLPAE QKGRSSHPCY GDTGVAYPQM ANTSAIAAAF GGGLPPGITG 240
241 TNDRCTVLVS NLNADSIDED KLFNLFSLYG NIVRIKLLRN KPDHALVQMG DGFQAELAVH 300
301 FLKGAMLFGK RLEVNFSKHP NITPGTDSHD YVNSNLNRFN RNAAKNYRYC CSPTKMIHLS 360
361 TLPQDVTEEE VMNHVQEHGA VVNTKVFEMN GKKQALVQFE NEEEAAEALV CKHATSLGGS 420
421 IIRISFSQLQ TI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
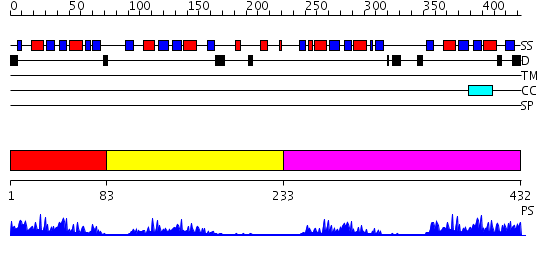
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 13.39794 | Solution structure of Polypyrimidine Tract Binding protein RBD1 complexed with CUCUCU RNA | |
| 2 | View Details | [83..232] | 13.522879 | NMR Structure of RRM2 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | |
| 3 | View Details | [233..432] | 36.0 | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)