













| General Information: |
|
| Name(s) found: |
AFG3L2
[HGNC (HUGO)]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 797 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAHRCLRLWG RGGCWPRGLQ QLLVPGGVGP GEQPCLRTLY RFVTTQARAS RNSLLTDIIA 60
61 AYQRFCSRPP KGFEKYFPNG KNGKKASEPK EVMGEKKESK PAATTRSSGG GGGGGGKRGG 120
121 KKDDSHWWSR FQKGDIPWDD KDFRMFFLWT ALFWGGVMFY LLLKRSGREI TWKDFVNNYL 180
181 SKGVVDRLEV VNKRFVRVTF TPGKTPVDGQ YVWFNIGSVD TFERNLETLQ QELGIEGENR 240
241 VPVVYIAESD GSFLLSMLPT VLIIAFLLYT IRRGPAGIGR TGRGMGGLFS VGETTAKVLK 300
301 DEIDVKFKDV AGCEEAKLEI MEFVNFLKNP KQYQDLGAKI PKGAILTGPP GTGKTLLAKA 360
361 TAGEANVPFI TVSGSEFLEM FVGVGPARVR DLFALARKNA PCILFIDEID AVGRKRGRGN 420
421 FGGQSEQENT LNQLLVEMDG FNTTTNVVIL AGTNRPDILD PALLRPGRFD RQIFIGPPDI 480
481 KGRASIFKVH LRPLKLDSTL EKDKLARKLA SLTPGFSGAD VANVCNEAAL IAARHLSDSI 540
541 NQKHFEQAIE RVIGGLEKKT QVLQPEEKKT VAYHEAGHAV AGWYLEHADP LLKVSIIPRG 600
601 KGLGYAQYLP KEQYLYTKEQ LLDRMCMTLG GRVSEEIFFG RITTGAQDDL RKVTQSAYAQ 660
661 IVQFGMNEKV GQISFDLPRQ GDMVLEKPYS EATARLIDDE VRILINDAYK RTVALLTEKK 720
721 ADVEKVALLL LEKEVLDKND MVELLGPRPF AEKSTYEEFV EGTGSLDEDT SLPEGLKDWN 780
781 KEREKEKEEP PGEKVAN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
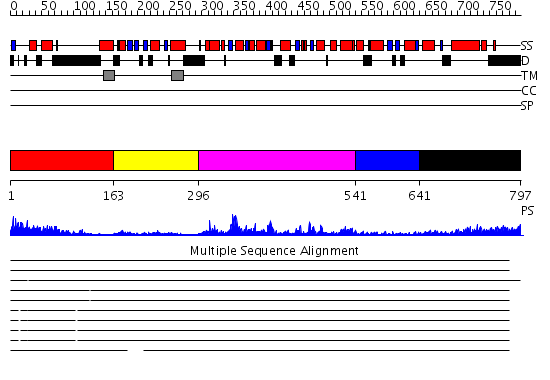
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | 0.993 | 30S subunit | |
| 2 | View Details | [163..295] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [296..540] | 64.39794 | AAA domain of cell division protein FtsH | |
| 4 | View Details | [541..640] | 4.47 | No description for 2di4A was found. | |
| 5 | View Details | [641..797] | 86.69897 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|