













| General Information: |
|
| Name(s) found: |
gi|41619044
[NCBI NR]
gi|21700907 [NCBI NR] gi|18410146 [NCBI NR] gi|15010654 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 624 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to salt stress
[IEP]
response to cadmium ion [IEP] |
| Molecular Function: |
DNA binding
[IEA][ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTVVGTVED RRDMFEGNAQ TRITPFSSTN QIGNPVAYKL VRVSGDGSLV PATDEEILEV 60
61 NDTDMHIPSD TCQTIGYLAT DEENVEVDET DMHIASDACQ TIGYLPAEGI PSRLSQIESS 120
121 EAINSGLLHS DNVQPYTDQY NEEMLQKVEQ EERLENVHGS QMPSTPADAN IQCSNENNFF 180
181 EEDQVHHEAL LQDECKMNES DMMERCSNAV ASPKETALSA AAQKPDFSRV RGEICLDNLP 240
241 IKALQETFRA TFGRDTTVKD KTWLKRRIAM GLINSCDVPT TNLRVKDNKL IGNQEKSNDV 300
301 TNAIRKEMGD DVRATKMKDA PSSTDHVNGH SNGGNHYYAS EDYSSEQRAA KRVRKPTRRY 360
361 IEELSETDDK QQNDKSVIPS KDQRLSEKSE VRSISVSSGK RVTVTRMVSL AGSEIEVPYV 420
421 SHVRRSRPRE NIMALLGCHS SYLEDKASAA ESNLNLSPSQ LSSEVVNRDS VEKSASRPVQ 480
481 NEFATSDENN VEHILSEVDQ EMEPEHIDSS GNSSDENNIG VPIMQGGALR RKHHRAWTLS 540
541 EIAKLVEGVS KYGAGKWSEI KKHLFSSHSY RTSVDLKDKW RNLLKTSFAQ SPSNSVGSLK 600
601 KHGSMHIPTQ ILLRVRELAE KQSQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
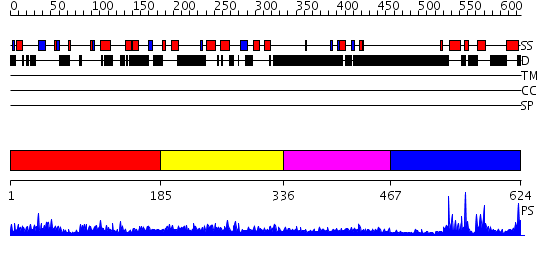
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [185..335] | 31.69897 | TRF2 | |
| 3 | View Details | [336..466] | 2.130994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [467..624] | 28.522879 | c-Myb, DNA-binding domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)