













| General Information: |
|
| Name(s) found: |
gi|28827394
[NCBI NR]
gi|28393068 [NCBI NR] gi|15230899 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 372 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
rRNA processing
[IEA]
|
| Molecular Function: |
RNA methyltransferase activity
[IEA]
catalytic activity [IEA] iron-sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLKSVFDAS EIKSEFESAG INPKFAIQIW KYVIQNPDCV WDEIPSLPSA AYSLLHSKFK 60
61 TLTSSLHSLF HSSDGTTSKL LIKLQNGAFV EAVVMRYDTR LGMLGGKPRP GGIRSTLCIS 120
121 SQVGCKMGCT FCATGTMGFK SNLTSGEIVE QLVHASRIAD IRNIVFMGMG EPLNNYNAVV 180
181 EAVRVMLNQP FQLSPKRITI STVGIVHAIN KLHNDLPGVS LAVSLHAPVQ EIRCQIMPAA 240
241 RAFPLQKLMD ALQTFQKNSQ QKIFIEYIML DGVNDQEQHA HLLGELLKTF QVVINLIPFN 300
301 PIGSTSQFET SSIQGVSRFQ KILRETYKIR TTIRKEMGQD ISGACGQLVV NQPDIKKTPG 360
361 TVELRDIEDL LL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
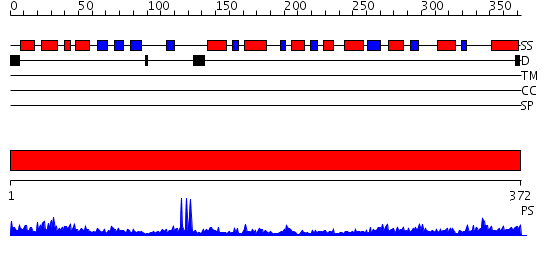
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..372] | 39.221849 | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)