













| General Information: |
|
| Name(s) found: |
DRP1E_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 624 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
vacuole [IDA] plasma membrane [IDA] |
| Biological Process: |
vesicle-mediated transport
[ISS]
response to cadmium ion [IEP] defense response to fungus [IMP] |
| Molecular Function: |
GTP binding
[IEA][ISS]
GTPase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTMESLIGL VNRIQRACTV LGDYGGGTGS NAFNSLWEAL PTVAVVGGQS SGKSSVLESI 60
61 VGRDFLPRGS GIVTRRPLVL QLHKTDDGTE EYAEFLHLPK KQFTDFALVR REIQDETDRI 120
121 TGKNKQISPV PIHLSIYSPN VVNLTLIDLP GLTKVAVEGQ PETIAEDIES MVRTYVDKPN 180
181 CIILAISPAN QDIATSDAIK LAKDVDPTGE RTFGVLTKLD LMDKGTNALE VLEGRSYRLQ 240
241 HPWVGIVNRS QADINKNVDM MLARRKEREY FDTSPDYGHL ASKMGSEYLA KLLSKHLESV 300
301 IRTRIPSILS LINKSIEELE RELDRMGRPV AVDAGAQLYT ILEMCRAFDK IFKEHLDGGR 360
361 PGGDRIYGVF DNQLPAALKK LPFDRHLSLQ SVKKIVSEAD GYQPHLIAPE QGYRRLIEGA 420
421 LGYFRGPAEA SVDAVHYVLK ELVRKSISET EELKRFPSLQ VELAAAANSS LEKFREESKK 480
481 SVIRLVDMES AYLTAEFFRK LPQEIERPVT NSKNQTASPS SATLDQYGDG HFRRIASNVS 540
541 AYVNMVSDTL RNTIPKACVY CQVRQAKLAL LNYFYSQISK REGKQLGQLL DEDPALMDRR 600
601 LECAKRLELY KKARDEIDAV AWVR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
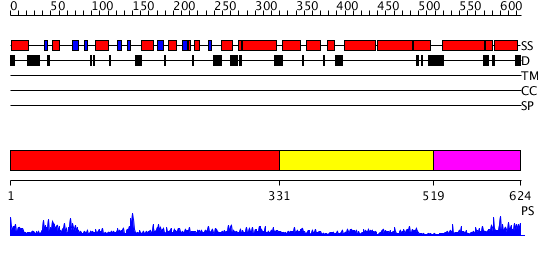
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..330] | 89.045757 | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | |
| 2 | View Details | [331..518] | 10.318759 | No description for PF01031.11 was found. No confident structure predictions are available. | |
| 3 | View Details | [519..624] | 35.431798 | No description for PF02212.9 was found. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)