













| General Information: |
|
| Name(s) found: |
RLM3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 796 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intrinsic to membrane
[IEA]
|
| Biological Process: |
defense response to fungus, incompatible interaction
[IMP]
[NOT]jasmonic acid and ethylene-dependent systemic resistance [IMP] defense response [ISS] |
| Molecular Function: |
ATP binding
[IEA]
transmembrane receptor activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSSSSQSYD VFPNFRGEDV RHSLVSHLRK ELDRKFINTF NDNRIERSRK ITPELLLAIE 60
61 NSRISLVVFS KNYASSTWCL DELVKIQECY EKLDQMVIPI FYKVDPSHVR KQTGEFGMVF 120
121 GETCKGRTEN EKRKWMRALA EVAHLAGEDL RNWRSEAEML ENIAKDVSNK LFPPSNNFSD 180
181 FVGIEAHIEA LISMLRFDSK KARMIGICGP SETGKTTIGR ALYSRLKSDF HHRAFVAYKR 240
241 KIRSDYDQKL YWEEQFLSEI LCQKDIKIEE CGAVEQRLKH TKVLIVLDDV DDIELLKTLV 300
301 GRIRWFGSES KIVVITQKRE LLKAHNIAHV YEVGFPSEEL AHQMFCRYAF GKNSPPHGFN 360
361 ELADEAAKIA GNRPKALKYV GSSFRRLDKE QWVKMLSEFR SNGNKLKISY DELDGKGQDY 420
421 VACLTNGSNS QVKAEWIHLA LGVSILLNIR SDGTTILKHL SYNRSMAQQA KIWWYENLER 480
481 VCKKYNICGI DSSTDGGGST YGQCSNSQFQ RNMDASPGGN KTSNQSTKDS PRASQVEKEK 540
541 IEYCEPHVYI TPAIFSDGTR APKYVESSSR RVTQVHHAKT WWPENCEKVY ENHNNIYGID 600
601 RSIDGGDKFE GKSKVSDGGL DGKDQGSMYG QSSNSELQIN MDADNRRCEP VSEMLFKNYN 660
661 VCSPNGLTDV NCSNPQSQRK LDASLKKDKI VHEWIRTGSG FFFDFQGPKS IVSAAQVDEK 720
721 NFEYCEQGVY ITLGILSGGI IVLKHLEFSR RMAQQAKVWW SENWIKVYQE HNICGIDKSF 780
781 DGRFDDRRVI RQLRPN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
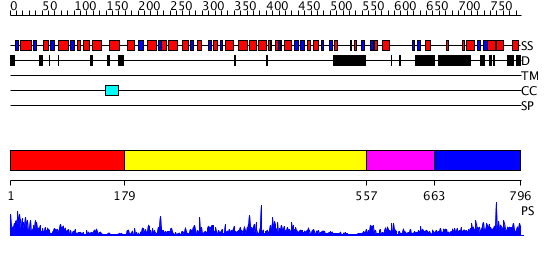
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 4.34 | Toll-like receptor 1, TLR1 | |
| 2 | View Details | [179..556] | 18.30103 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [557..662] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [663..796] | 6.76998 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)