













| General Information: |
|
| Name(s) found: |
UBP13_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1115 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEA]
|
| Molecular Function: |
ubiquitin-specific protease activity
[ISS]
ubiquitin thiolesterase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTMMTPPPLD QQEDEEMLVP NPDLVEGPQP MEVAQTDPAA TAVENPPPED PPSLKFTWTI 60
61 PMFTRLNTRK HYSDVFVVGG YKWRILIFPK GNNVDHLSMY LDVADAANLP YGWSRYSQFS 120
121 LAVVNQVNNR YSIRKETQHQ FNARESDWGF TSFMPLSELY EPTRGYLVND TVLIEAEVAV 180
181 RKVLDYWSYD SKKETGFVGL KNQGATCYMN SLLQTLYHIP YFRKAVYHMP TTENDAPTAS 240
241 IPLALQSLFY KLQYNDTSVA TKELTKSFGW DTYDSFMQHD VQELNRVLCE KLEDKMKGTV 300
301 VEGTIQKLFE GHHMNYIECI NVDYKSTRKE SFYDLQLDVK GCKDVYASFD KYVEVERLEG 360
361 DNKYHAEGHD LQDAKKGVLF IDFPPVLQLQ LKRFEYDFMR DTMVKINDRY EFPLQLDLDR 420
421 EDGRYLSPDA DKSVRNLYTL HSVLVHSGGV HGGHYYAFIR PTLSDQWYKF DDERVTKEDV 480
481 KRALEEQYGG EEELPQNNPG FNNPPFKFTK YSNAYMLVYI RESDKDKIIC NVDEKDIAEH 540
541 LRVRLKKEQE EKEDKRKYKA QAHLFTTIKV ARDDDITEQI GKNIYFDLVD HEKVRSFRIQ 600
601 KQTPFQQFKE EVAKEFGVPV QLQRFWIWAK RQNHTYRPNR PLSPNEELQT VGQIREASNK 660
661 ANNAELKLFL EIERGPDDLP IPPPEKTSED ILLFFKLYDP ENAVLRYVGR LMVKSSSKPM 720
721 DIVGQLNKMA GFAPDEEIEL FEEIKFEPCV MCEQIDKKTS FRLCQIEDGD IICYQKPLSI 780
781 EESEFRYPDV PSFLEYVQNR ELVRFRTLEK PKEDEFTMEL SKLHTYDDVV ERVAEKLGLD 840
841 DPSKLRLTSH NCYSQQPKPQ PIKYRGVDHL SDMLVHYNQT SDILYYEVLD IPLPELQGLK 900
901 TLKVAFHSAT KDEVIIHNIR LPKQSTVGDV INELKTKVEL SHQDAELRLL EVFFHKIYKI 960
961 FPSTERIENI NDQYWTLRAE EIPEEEKNIG PNDRLIHVYH FTKEAGQNQQ VQNFGEPFFL1020
1021 VIHEGETLEE IKTRIQKKLH VPDEDFAKWK FASFSMGRPD YLLDTDVVYN RFQRRDVYGA1080
1081 WEQYLGLEHI DNAPKRAYAA NQNRHAYEKP VKIYN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
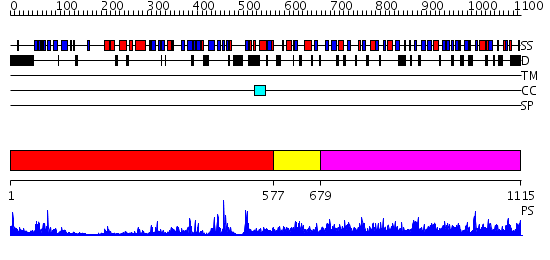
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..576] | 84.39794 | Crystal structure of HAUSP | |
| 2 | View Details | [577..678] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [679..1115] | 1.34 | No description for 2ibiA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)