













| General Information: |
|
| Name(s) found: |
AT18G_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 959 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to starvation
[IEP]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMKKGKGKNS GLLPNSFKII SSCLKTVSAN ATNVASSVRS AGASVAASIS AAEDDKDQVT 60
61 WAGFGILELG QHVTRHVLLL GYQNGFQVFD VEDASNFNEL VSKRGGPVSF LQMQPLPARS 120
121 GDHEGFWNSH PLLLVVAGDE TNGTGLGHSF SQNGSLARDG SSDSKAGDAI NYPTTVRFYS 180
181 LRSHSYVYVL RFRSSVCMIR CSSRVVAVGL ANQIYCVDAL TLENKFSVLT YPVPQPVRQG 240
241 TTRVNVGYGP MAVGPRWLAY ASKSSMTMKT GRLSPQTFTS SPSLSPSSSS GGSSFMARYA 300
301 MESSKQLANG LINLGDMGYK TLSKYCQDML PDGSTSPASP NAIWKVGGVS GSDAENAGMV 360
361 AVKDLVSGAL VSQFKAHTSP ISALCFDPSG TLLVTASVCG NNINVFQIMP SRSHNAPGDL 420
421 SYEWESSHVH LFKLHRGITS AIVQDICFSQ QSQWVAIISS KGTCHIFVLN SSGSDAAFQP 480
481 CEGEEPTRLP ASSLPWWFTQ SLSSNQQSLS PPTAVALSVV SRIKYSSFGW LNTVSNATTA 540
541 ATGKVFVPSG AVAAVFHKSV THDLQLNSRT NALEHILVYT PSGHVVQHEL LPSVCTESPE 600
601 NGLRVQKTSH VQVQEDDLRV KVEPIQWWDV CRRSDWLETE ERLPKSITEK QYDLETVSNH 660
661 LTSHEDACLS LDMNSHFSED KYLKSCSEKP PERSHCYLSN FEVKVTSGML PVWQNSKISF 720
721 HVMDSPRDSS STGGEFEIEK VPAHELEIKQ KKLLPVFDHF HSTKATLEDR FSMKCYHTSA 780
781 TGSHQVNGKI CQDIINCHSK PGSIESAESS EEGSTKQMEN LHDSDHMSNS IKSSLPLYPT 840
841 VNGIYKEIEK NNANGWMEKP VTAKLSTLKE TRITNGFTTP PILTDSVNEQ MLSTGKPPMG 900
901 FGFALHEEHC KAVADPKEEH LKKKLDEVTN VHHLNVNNNN TEKLQGDKMV HGMVSFVGD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
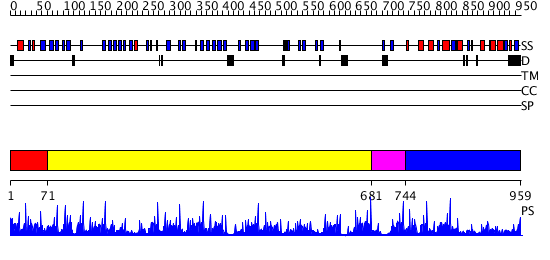
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | 6.69897 | Surface layer protein | |
| 2 | View Details | [71..680] | 34.0 | Actin interacting protein 1 | |
| 3 | View Details | [681..743] | 59.39794 | Actin interacting protein 1 | |
| 4 | View Details | [744..959] | 47.045757 | Homology Model of Yeast RACK1 Protein fitted into 11.7A cryo-EM map of Yeast 80S Ribosome |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.52 |
Source: Reynolds et al. (2008)