













| General Information: |
|
| Name(s) found: |
CMTA3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1032 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to biotic stimulus
[IMP]
|
| Molecular Function: |
calmodulin binding
[IEP][TAS]
transcription regulator activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEARRFSPV HELDVGQILS EARHRWLRPP EICEILQNYQ RFQISTEPPT TPSSGSVFMF 60
61 DRKVLRYFRK DGHNWRKKKD GKTVKEAHER LKAGSVDVLH CYYAHGQDNE NFQRRSYWLL 120
121 QEELSHIVFV HYLEVKGSRV STSFNRMQRT EDAARSPQET GDALTSEHDG YASCSFNQND 180
181 HSNHSQTTDS ASVNGFHSPE LEDAESAYNQ HGSSTAYSHQ ELQQPATGGN LTGFDPYYQI 240
241 SLTPRDSYQK ELRTIPVTDS SIMVDKSKTI NSPGVTNGLK NRKSIDSQTW EEILGNCGSG 300
301 VEALPLQPNS EHEVLDQILE SSFTMQDFAS LQESMVKSQN QELNSGLTSD RTVWFQGQDM 360
361 ELNAISNLAS NEKAPYLSTM KQHLLHGALG EEGLKKMDSF NRWMSKELGD VGVIADANES 420
421 FTQSSSRTYW EEVESEDGSN GHNSRRDMDG YVMSPSLSKE QLFSINDFSP SWAYVGCEVV 480
481 VFVTGKFLKT REETEIGEWS CMFGQTEVPA DVISNGILQC VAPMHEAGRV PFYVTCSNRL 540
541 ACSEVREFEY KVAESQVFDR EADDESTIDI LEARFVKLLC SKSENTSPVS GNDSDLSQLS 600
601 EKISLLLFEN DDQLDQMLMN EISQENMKNN LLQEFLKESL HSWLLQKIAE GGKGPSVLDE 660
661 GGQGVLHFAA SLGYNWALEP TIIAGVSVDF RDVNGWTALH WAAFFGRERI IGSLIALGAA 720
721 PGTLTDPNPD FPSGSTPSDL AYANGHKGIA GYLSEYALRA HVSLLSLNDK NAETVEMAPS 780
781 PSSSSLTDSL TAVRNATQAA ARIHQVFRAQ SFQKKQLKEF GDKKLGMSEE RALSMLAPKT 840
841 HKSGRAHSDD SVQAAAIRIQ NKFRGYKGRK DYLITRQRII KIQAHVRGYQ FRKNYRKIIW 900
901 SVGVLEKVIL RWRRKGAGLR GFKSEALVEK MQDGTEKEED DDFFKQGRKQ TEDRLQKALA 960
961 RVKSMVQYPE ARDQYRRLLN VVNDIQESKV EKALENSEAT CFDDDDDLID IEALLEDDDT1020
1021 LMLPMSSSLW TS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
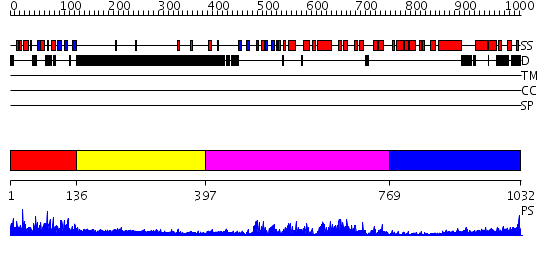
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 88.420216 | No description for PF03859.7 was found. No confident structure predictions are available. | |
| 2 | View Details | [136..396] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [397..768] | 68.30103 | Ankyrin-R | |
| 4 | View Details | [769..1032] | 14.39794 | No description for 2dfsA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)