













| General Information: |
|
| Name(s) found: |
PEX6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 941 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein import into peroxisome matrix
[IMP]
fatty acid beta-oxidation [IMP] |
| Molecular Function: |
ATPase activity
[ISS]
nucleotide binding [IEA] ATP binding [IEA] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVERRNPLVL SSTRSTLRSV LNSSQPSSAD GDRVLNKDGD LLRGNARLSA GILRWRKDGE 60
61 NVSDAKLDSL DDSALVGLST QLLKRLSINS GSLVVVKNIE IGIQRVAQVV VLDPPKTTLE 120
121 DASLTQVPVS DSLHTMLVFP TYDLMGQQLL DQEVAYLSPM LAFNLSLHIS CLKSLVHRGN 180
181 GVLEKYFEAK CDEEFIGKSA EDGSKIGLDL EPVSQVPGYA SHLRVSFVKI PECGTIPSLK 240
241 VNSSFEAEER QGLIDSALQK YFGTDRQLSR GDIFRIYIDW NCGSSICNPC SQRLCSESDD 300
301 YIYFKVIAME PSNERFLRVN HSQTALVLGG TVSSGLPPDL LVYRSKVPMP LQEETVNILA 360
361 SVLSPPLCPS ALASKLRVAV LLHGIPGCGK RTVVKYVARR LGLHVVEFSC HSLLASSERK 420
421 TSTALAQTFN MARRYSPTIL LLRHFDVFKN LGSQDGSLGD RVGVSFEIAS VIRELTEPVS 480
481 NGDSSMEEKS NSNFSENEVG KFRGHQVLLI ASAESTEGIS PTIRRCFSHE IRMGSLNDEQ 540
541 RSEMLSQSLQ GVSQFLNISS DEFMKGLVGQ TSGFLPRDLQ ALVADAGANL YISQESETKK 600
601 INSLSDDLHG VDIHQASQID NSTEKLTAKE DFTKALDRSK KRNASALGAP KVPNVKWDDV 660
661 GGLEDVKTSI LDTVQLPLLH KDLFSSGLRK RSGVLLYGPP GTGKTLLAKA VATECSLNFL 720
721 SVKGPELINM YIGESEKNVR DIFEKARSAR PCVIFFDELD SLAPARGASG DSGGVMDRVV 780
781 SQMLAEIDGL SDSSQDLFII GASNRPDLID PALLRPGRFD KLLYVGVNAD ASYRERVLKA 840
841 LTRKFKLSED VSLYSVAKKC PSTFTGADMY ALCADAWFQA AKRKVSKSDS GDMPTEEDDP 900
901 DSVVVEYVDF IKAMDQLSPS LSITELKKYE MLRDQFQGRS S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
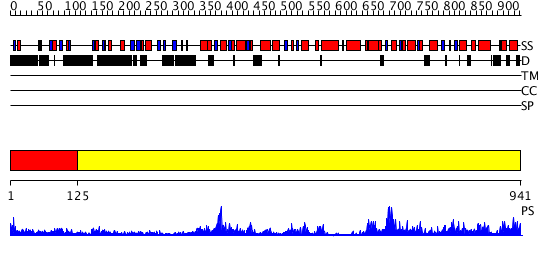
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [125..941] | 157.0 | No description for 3cf1A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)