













| General Information: |
|
| Name(s) found: |
gi|190404562
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 772 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDINSNASVS PRPDGLPMTA GYNSASGKVR NSIRSIINHP EDSARAKERS ETNSPKNNGN 60
61 KKPRKKRKTF SCDTCRRVKT RCDFEPFIGK CYRCNVLQLD CSLARNKDNE ILNTLREDGL 120
121 LKKINSINHN LGSFSHLNAD SPNESQSSFE KNGTVNFDNY MIDKRLSSLE EHIKSLHQKM 180
181 DLIITTAKMS YNSDIKGPGD DIQNVDFSSN KTYDSRLTSG SETIRKTGEY RKENLFLNGF 240
241 KLKESPLKLL HDIDERLFPS KATSKAAKLA GQQRPYAVAR VNFLHFYENN QELCHKLAKE 300
301 FLVRSHFWII PGGRKEIDVE YAHSHLFITS VFTIIAMSFA DNDKYAAEQE ILYPLVERLL 360
361 TNTLTMFEKL TAFDIEAILY CCMFHISRKA KRYRQLKFNS LVLSNFALNS LLHVIDFYQI 420
421 KDRVLVKEVY NPEDLYHLRI LNSLTACYLE YSISYGDIRE QDDMLKEFNK LVAKFPQANF 480
481 GDDIKISEIN LGDIVNGIFI NLKNYFAQCL DDFNNDRYGG NADTFIFVFP ELNYWLKNWE 540
541 ELLAKDGAGV LLFTFDFYHI MICRTFITEF SSTLKSNQRF LKLILNTMKE HSFSLLNGFL 600
601 RLPPTLIRGA PIFTCHQLVY ACLTLCDYLY WFDSSERQRV LSLCTKVYWH LSTIGEKMNE 660
661 ATDNVGKIIK SIIDTSKTRI NFGSLSKENS DNDKMSTNAN NYTGAGNLHA AKPATSPTNV 720
721 GTLHENLSSS HFMIPDVDQF NSFEDFFQDF FDSLKPNSQK MFTSDKKTEQ TT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
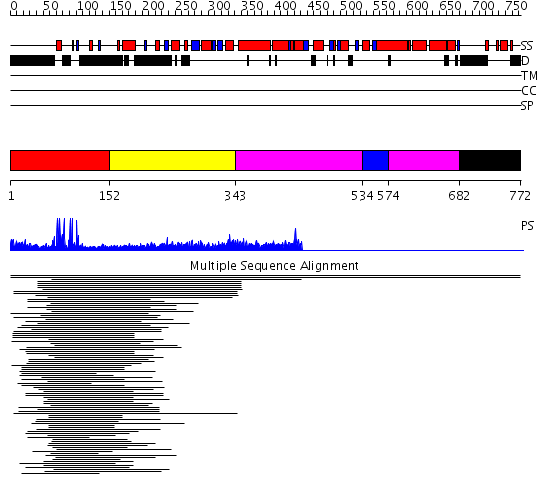
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 12.09691 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [152..342] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [343..533] [574..681] |
7.33 | Hemolysin E (HlyE, ClyA, SheA) | |
| 4 | View Details | [534..573] | 7.33 | Hemolysin E (HlyE, ClyA, SheA) | |
| 5 | View Details | [682..772] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)