













| General Information: |
|
| Name(s) found: |
gi|187252543
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 754 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTAGRGNLG LIPRSTAFQK QEGRLTVKQE PANQTWGQGS SLQKNYPPVC EIFRLHFRQL 60
61 CYHEMSGPQE ALSRLRELCR WWLMPEVHTK EQILELLVLE QFLSILPGEL RTWVQLHHPE 120
121 SGEEAVAVVE DFQRHLSGSE EVSAPAQKQE MHFEETTALG TTKESPPTSP LSGGSAPGAH 180
181 LEPPYDPGTH HLPSGDFAQC TSPVPTLPQV GNSGDQAGAT VLRMVRPQDT VAYEDLSVDY 240
241 TQKKWKSLTL SQRALQWNMM PENHHSMASL AGENMMKGSE LTPKQEFFKG SESSNRTSGG 300
301 LFGVVPGAAE TGDVCEDTFK ELEGQTSDEE GSRLENDFLE ITDEDKKKST KDRYDKYKEV 360
361 GEHPPLSSSP VEHEGVLKGQ KSYRCDECGK AFNRSSHLIG HQRIHTGEKP YECNECGKTF 420
421 RQTSQLIVHL RTHTGEKPYE CSECGKAYRH SSHLIQHQRL HNGEKPYKCN ECAKAFTQSS 480
481 RLTDHQRTHT GEKPYECNEC GEAFIRSKSL ARHQVLHTGK KPYKCNECGR AFCSNRNLID 540
541 HQRIHTGEKP YECSECGKAF SRSKCLIRHQ SLHTGEKPYK CSECGKAFNQ NSQLIEHERI 600
601 HTGEKPFECS ECGKAFGLSK CLIRHQRLHT GEKPYKCNEC GKSFNQNSHL IIHQRIHTGE 660
661 KPYECNECGK VFSYSSSLMV HQRTHTGEKP YKCNDCGKAF SDSSQLIVHQ RVHTGEKPYE 720
721 CSECGKAFSQ RSTFNHHQRT HTGEKSSGLA WSVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
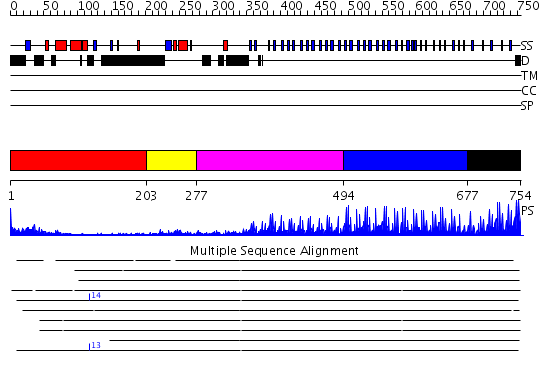
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] | 4.07 | Solution structure of the SCAN homodimer from MZF-1/ZNF42 | |
| 2 | View Details | [203..276] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..493] | 7.9 | No description for 2i13A was found. | |
| 4 | View Details | [494..676] | 40.30103 | No description for 2i13A was found. | |
| 5 | View Details | [677..754] | 6.0 | ZIF268 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)