













| General Information: |
|
| Name(s) found: |
gi|220952208
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 533 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPFPSLQECE QMVQMLRVVE LQKILSFLNI SFAGRKTDLQ SRILSFLRTN LELLAPKVQE 60
61 VYAQSVQEQN ATLQYIDPTR MYSHIQLPPT VQPNPVGLVG SGQGVQVPGG QMNVVGGAPF 120
121 LHTHSINSQL PIHPDVRLKK LAFYDVLGTL IKPSTLVPRN TQRVQEVPFY FTLTPQQATE 180
181 IASNRDIRNS SKVEHAIQVQ LRFCLVETSC DQEDCFPPNV NVKVNNKLCQ LPNVIPTNRP 240
241 NVEPKRPPRP VNVTSNVKLS PTVTNTITVQ WCPDYTRSYC LAVYLVKKLT STQLLQRMKT 300
301 KGVKPADYTR GLIKEKLTED ADCEIATTML KVSLNCPLGK MKMLLPCRAS TCSHLQCFDA 360
361 SLYLQMNERK PTWNCPVCDK PAIYDNLVID GYFQEVLGSS LLKSDDTEIQ LHQDGSWSTP 420
421 GLRSETQILD TPSKPAQKVE VISDDIELIS DDAKPVKRDL SPAQDEQPTS TSNSETVDLT 480
481 LSDSDDDMPL AKRRPPAKQA VASSTSNGSG GGQRAYTPAQ QPQQSGTLDP FLQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
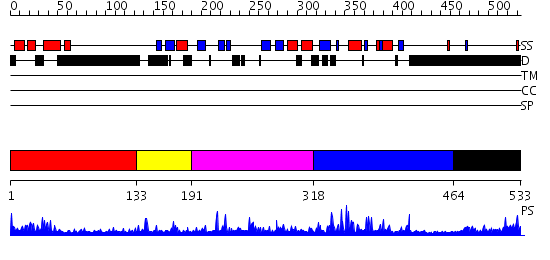
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 15.69897 | Solution structure of human p53 binding domain of PIAS-1 | |
| 2 | View Details | [133..190] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [191..317] | 2.131984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [318..463] | 10.69897 | No description for 2yu4A was found. | |
| 5 | View Details | [464..533] | 1.095995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)