













| General Information: |
|
| Name(s) found: |
gi|195566518
[NCBI NR]
gi|194204219 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Drosophila simulans |
| Length: | 515 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQRFAPGQA PVQSRYQPPP PAPGMRPYPP PGASFPPRGF PLHPNTTQPV PGTANVAGVP 60
61 GVPGVPGVPG PSLLQRAAQQ PGFQSSLRGT GAGGGGVGSG GGSKRSNESR SLGGGGSKSD 120
121 FATAKKKKKL AEKILPQKVR DLVPESQAYM DLLTFERKLD ATIMRKRLDI QEALKRPMKQ 180
181 KRKLRIFISN TFYPSKEPTN DGEEGAVASW ELRVEGRLLE DGKGDPNTKI KRKFSSFFKS 240
241 LVIELDKELY GPDNHLVEWH RTHTTQETDG FQVKRPGDRN VRCTILLLLD YQPLQFKLDP 300
301 RLARLLGVHT QTRPVIISAL WQYIKTHKLQ DAHEREYINC DKYLEQIFSC QRMKFAEIPQ 360
361 RLNPLLHPPD PIVINHFIES GAENKQTACY DIDVEVDDTL KNQMNSFLMS TASQQEIQGL 420
421 DTKIHETVDT INQMKTNREF FLSFAKDPQM FIHRWIISET RDLKLMTDVA GNPEEERRAE 480
481 FYYQPWTHEA VSRYFFTKVN QKRAELEQAL GIRNG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
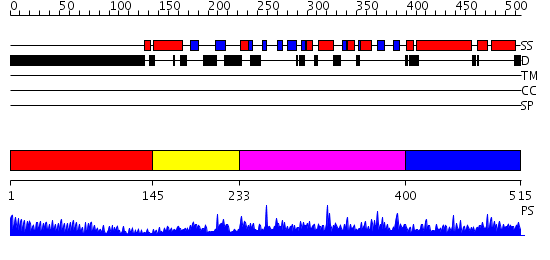
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 23.39794 | No description for 1y0fB was found. | |
| 2 | View Details | [145..232] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [233..399] | 30.69897 | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | |
| 4 | View Details | [400..515] | 1.058952 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)