













| General Information: |
|
| Name(s) found: |
gi|218197827
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Oryza sativa Indica Group |
| Length: | 711 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENGDKDEKN VAVTEGSTNS EEKDQDEDLL RRTEMLNAKE AINSSNENSG NDSEAQIHVR 60
61 DDSQKEFNEK MNKEGSSDPM EPTDSSQTEE DILAEDKSEE PVFDGTEVAE MEDLRRSSNQ 120
121 SVELDSDAHG SVLNERATAI KNFVKEKGAI AVSTFIRRLS GKKDENEFSV EDEKNEGSES 180
181 ISSGNIGSDA EPKSKEVQPK SEERTTWNPL NLIKIGRDFD TFMTGEAGHE NVPDLIEQPT 240
241 GKGRIIIYTK LGCEDCKMVR SFMRQKMLKY VEINIDIFPS RKMELENNTG SSTVPKVYFN 300
301 DLLIGGLTEL KKMEESGILD DRTDALFKDE PSSAAPLPPL PGEDDESGSG KIDELATIVR 360
361 KMRESITLKD RFYKMRRFSS CFLGSEAVDF LSEDQYLERD EAVEFGRKLA SKYFYRHVLD 420
421 EDVFEDGNHL YRFLDNDPII MSQCYNIPKG IIDVEPKPIV EVASRLRKLS ETMFEAYVSE 480
481 DGKHVDYRSI QGCEEFKRYV RTTEELQRVE THELSREEKL AFFINLYNMM AIHALVTCGH 540
541 PAGPLDRRKF FGDFKYVIGG CAYSMSAIQN GILRGNQRPP YNLAKPFGQK DQRSKVALPY 600
601 AEPLVHFALV CGTKSGPALR CYSPGNIDKE LVEAARDFLR NVGIVVDPEA KVASVSKILR 660
661 WYSTDFGKNE TEVLKHAANY LEPAESEQFL ELLANTQLKV LYQPYDWSLN I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
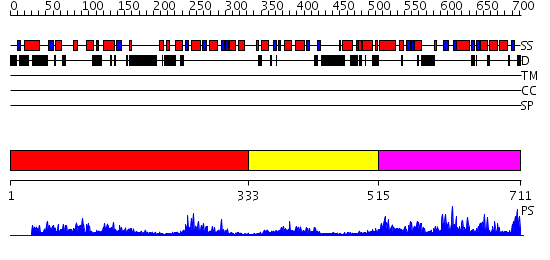
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..332] | 48.221849 | C-terminal, Grx domain of Hybrid-Prx5; N-terminal, Prx domain of Hybrid-Prx5 | |
| 2 | View Details | [333..514] | 17.30103 | Regulatory domain of epac2, domain 2; Regulatory domain of Epac2, domains 1 and 3 | |
| 3 | View Details | [515..711] | 3.148981 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)