













| General Information: |
|
| Name(s) found: |
PKL_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1384 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
chromatin modification
[ISS]
cell proliferation [IMP] root development [IMP] response to gibberellin stimulus [IMP] negative regulation of transcription [IMP] response to auxin stimulus [IMP] negative regulation of abscisic acid mediated signaling pathway [IMP] |
| Molecular Function: |
ATPase activity
[ISS]
DNA binding [ISS] DNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLVERLRI RSDRKPVYNL DDSDDDDFVP KKDRTFEQVE AIVRTDAKEN ACQACGESTN 60
61 LVSCNTCTYA FHAKCLVPPL KDASVENWRC PECVSPLNEI DKILDCEMRP TKSSEQGSSD 120
121 AEPKPIFVKQ YLVKWKGLSY LHCSWVPEKE FQKAYKSNHR LKTRVNNFHR QMESFNNSED 180
181 DFVAIRPEWT TVDRILACRE EDGELEYLVK YKELSYDECY WESESDISTF QNEIQRFKDV 240
241 NSRTRRSKDV DHKRNPRDFQ QFDHTPEFLK GLLHPYQLEG LNFLRFSWSK QTHVILADEM 300
301 GLGKTIQSIA LLASLFEENL IPHLVIAPLS TLRNWEREFA TWAPQMNVVM YFGTAQARAV 360
361 IREHEFYLSK DQKKIKKKKS GQISSESKQK RIKFDVLLTS YEMINLDSAV LKPIKWECMI 420
421 VDEGHRLKNK DSKLFSSLTQ YSSNHRILLT GTPLQNNLDE LFMLMHFLDA GKFGSLEEFQ 480
481 EEFKDINQEE QISRLHKMLA PHLLRRVKKD VMKDMPPKKE LILRVDLSSL QKEYYKAIFT 540
541 RNYQVLTKKG GAQISLNNIM MELRKVCCHP YMLEGVEPVI HDANEAFKQL LESCGKLQLL 600
601 DKMMVKLKEQ GHRVLIYTQF QHMLDLLEDY CTHKKWQYER IDGKVGGAER QIRIDRFNAK 660
661 NSNKFCFLLS TRAGGLGINL ATADTVIIYD SDWNPHADLQ AMARAHRLGQ TNKVMIYRLI 720
721 NRGTIEERMM QLTKKKMVLE HLVVGKLKTQ NINQEELDDI IRYGSKELFA SEDDEAGKSG 780
781 KIHYDDAAID KLLDRDLVEA EEVSVDDEEE NGFLKAFKVA NFEYIDENEA AALEAQRVAA 840
841 ESKSSAGNSD RASYWEELLK DKFELHQAEE LNALGKRKRS RKQLVSIEED DLAGLEDVSS 900
901 DGDESYEAES TDGEAAGQGV QTGRRPYRRK GRDNLEPTPL MEGEGRSFRV LGFNQSQRAI 960
961 FVQTLMRYGA GNFDWKEFVP RLKQKTFEEI NEYGILFLKH IAEEIDENSP TFSDGVPKEG1020
1021 LRIEDVLVRI ALLILVQEKV KFVEDHPGKP VFPSRILERF PGLRSGKIWK EEHDKIMIRA1080
1081 VLKHGYGRWQ AIVDDKELGI QELICKELNF PHISLSAAEQ AGLQGQNGSG GSNPGAQTNQ1140
1141 NPGSVITGNN NASADGAQVN SMFYYRDMQR RLVEFVKKRV LLLEKAMNYE YAEEYYGLGG1200
1201 SSSIPTEEPE AEPKIADTVG VSFIEVDDEM LDGLPKTDPI TSEEIMGAAV DNNQARVEIA1260
1261 QHYNQMCKLL DENARESVQA YVNNQPPSTK VNESFRALKS INGNINTILS ITSDQSKSHE1320
1321 DDTKPDLNNV EMKDTAEETK PLRGGVVDLN VVEGEENIAE ASGSVDVKME EAKEEEKPKN1380
1381 MVVD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
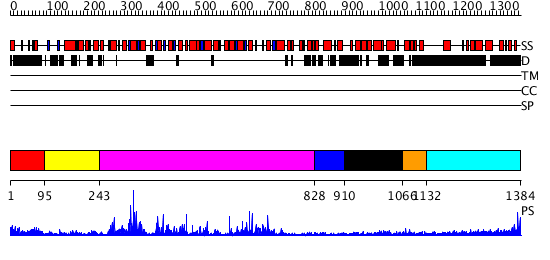
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [95..242] | 8.0 | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | |
| 3 | View Details | [243..827] | 83.69897 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 4 | View Details | [828..909] | 25.657577 | No description for PF06465.5 was found. No confident structure predictions are available. | |
| 5 | View Details | [910..1065] | 98.337242 | No description for PF06461.3 was found. No confident structure predictions are available. | |
| 6 | View Details | [1066..1131] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1132..1384] | 6.253997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)