













| General Information: |
|
| Name(s) found: |
gi|207340799
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae AWRI1631 |
| Length: | 565 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRTVISSSN AYASKRSRLD IEHDFEQYHS LNKKYYPRPI TRTGANQFNN KSRAKPMEIV 60
61 EKLQKKQKTS FENVSTVMHW FRNDLRLYDN VGLYKSVALF QQLRQKNAKA KLYAVYVINE 120
121 DDWRAHMDSG WKLMFIMGAL KNLQQSLAEL HIPLLLWEFH TPKSTLSNSK EFVEFFKEKC 180
181 MNVSSGTGTI ITANIEYQTD ELYRDIRLLE NEDHRLQLKY YHDSCIVAPG LITTDRGTNY 240
241 SVFTPWYKKW VLYVNNYKKS TSEICHLHII EPLKYNETFE LKPFQYSLPD EFLQYIPKSK 300
301 WCLPDVSEEA ALSRLKDFLG TKSSKYNNEK DMLYLGGTSG LSVYITTGRI STRLIVNQAF 360
361 QSCNGQIMSK ALKDNSSTQN FIKEVAWRDF YRHCMCNWPY TSMGMPYRLD TLDIKWENNP 420
421 VAFEKWCTGN TGIPIVDAIM RKLLYTGYIN NRSRMITASF LSKNLLIDWR WGERWFMKHL 480
481 IDGDSSSNVG GWGFCSSTGI DAQPYFRVFN MDIQAKKYDP QMIFVKQWVP ELISSENKRP 540
541 ENYPKPLVDL KHSRERALKV YKDAM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
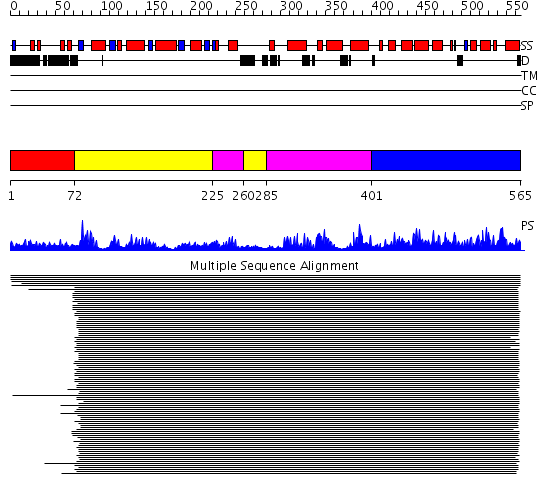
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [72..224] [260..284] |
809.0103 | C-terminal domain of DNA photolyase; DNA photolyase | |
| 3 | View Details | [225..259] [285..400] |
809.0103 | C-terminal domain of DNA photolyase; DNA photolyase | |
| 4 | View Details | [401..565] | 809.0103 | C-terminal domain of DNA photolyase; DNA photolyase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)