













| General Information: |
|
| Name(s) found: |
gi|190407122
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 897 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDIKNKDHT TSVNHNLMAS AGNYTAEKEI GKGSFATVYR GHLTSDKSQH VAIKEVSRAK 60
61 LKNKKLLENL EIEIAILKKI KHPHIVGLID CERTSTDFYL IMEYCALGDL TFLLKRRKEL 120
121 MENHPLLRTV FEKYPPPSEN HNGLHRAFVL SYLQQLASAL KFLRSKNLVH RDIKPQNLLL 180
181 STPLIGYHDS KSFHELGFVG IYNLPILKIA DFGFARFLPN TSLAETLCGS PLYMAPEILN 240
241 YQKYNAKADL WSVGTVVFEM CCGTPPFRAS NHLELFKKIK RANDVITFPS YCNIEPELKE 300
301 LICSLLTFDP AQRIGFEEFF ANKVVNEDLS SYELEDDLPE LESKSKGIVE SNMFVSEYLS 360
361 KQPKSPNSNL AGHQSMADNP AELSDALKNS NILTAPAVKT DHTQAVDKKA SNNKYHNSLV 420
421 SDRSFEREYV VVEKKSVEVN SLADEVAQAG FNPNPIKHPT STQNQNVLLN EQFSPNNQQY 480
481 FQNQGENPRL LRATSSSSGG SDGSRRPSLV DRRLSISSLN PSNALSRALG IASTRLFGGA 540
541 NQQQQQQQIT SSPPYSQTLL NSQLFHELTE NIILRIDHLQ HPETLKLDNT NIVSILESLA 600
601 AKAFVVYSYA EVKFSQIVPL STTLKGMANF ENRRSMDSNA IAEEQDSDDA EEEDETLKKY 660
661 KEDCLSTKTF GKGRTLSATS QLSATFNKLP RSEMILLCNE AIVLYMKALS ILSKSMQVTS 720
721 NWWYESQEKS CSLRVNVLVQ WLREKFNECL EKADFLRLKI NDLRFKHASE VAENQTLEEK 780
781 GSSEEPVYLE KLLYDRALEI SKMAAHMELK GENLYNCELA YATSLWMLET SLDDDDFTNA 840
841 YGDYPFKTNI HLKSNDVEDK EKYHSVLDEN DRIIIRKYID SIANRLKILR QKMNHQN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
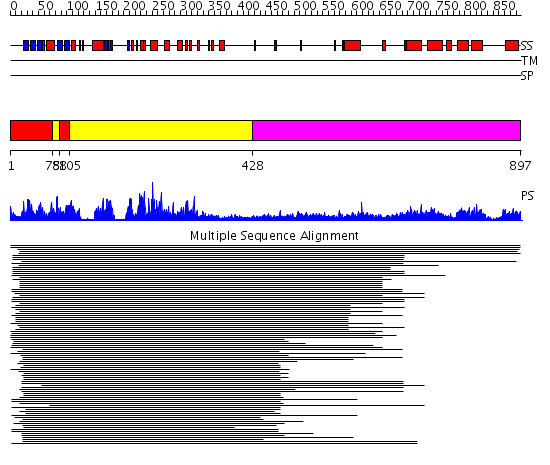
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] [88..104] |
384.9794 | cAMP-dependent PK, catalytic subunit | |
| 2 | View Details | [75..87] [105..427] |
384.9794 | cAMP-dependent PK, catalytic subunit | |
| 3 | View Details | [428..897] | 1.481956 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [678..897] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)